
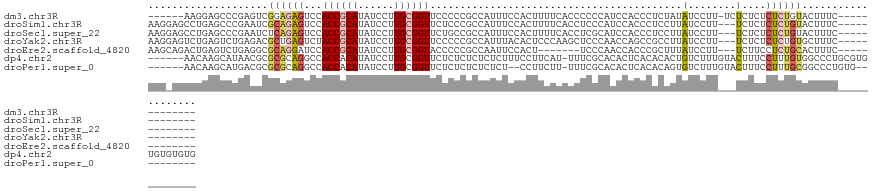
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,943,810 – 23,943,918 |
| Length | 108 |
| Max. P | 0.783204 |

| Location | 23,943,810 – 23,943,918 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 128 |
| Reading direction | reverse |
| Mean pairwise identity | 64.50 |
| Shannon entropy | 0.63157 |
| G+C content | 0.53327 |
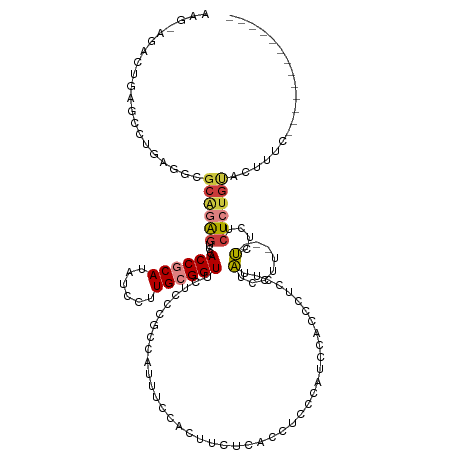
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -7.28 |
| Energy contribution | -8.26 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.783204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23943810 108 - 27905053 ------AAGGAGCCCGAGUCGGAGAGUCCACCGCAUAUCCUUGCGGUUCCCCCGCCAUUUCCACUUUUCACCCCCCAUCCACCCUCUAUAUCCUU-UCUCUCUCUCUGUACUUUC------------- ------.........(((((((((((...((((((......))))))......(.......).................................-.....))))))).))))..------------- ( -18.50, z-score = -1.42, R) >droSim1.chr3R 23643562 112 - 27517382 AAGGAGCCUGAGCCCGAAUCGCAGAGUCCACCGCAUAUCCUUGCGGUUCUCCCGCCAUUUCCACUUUUCACCUCCCAUCCACCCUCCUUAUCCUU---UCUCUCUCUGUACUUUC------------- ..((((...(((...((((.((.(((...((((((......)))))).)))..)).))))...))).....))))....................---.................------------- ( -21.00, z-score = -2.66, R) >droSec1.super_22 562529 112 - 1066717 AAGGAGCCUGAGCCCGAAUCUCAGAGUCCACCGCAUAUCCUUGCGGUUCUGCCGCCAUUUCCACUUUUCACCUCGCAUCCACCCUCCUUAUCCUU---UCUCUCUCUGUACUUUC------------- ((((((.(((((.......)))))((...((((((......)))))).)).................................))))))......---.................------------- ( -20.90, z-score = -1.91, R) >droYak2.chr3R 6280344 112 + 28832112 AAGGAGUCUGAGUCUGAGACGCUGAGUCUACCGCAUAUCCUUGCGGUUCCCCCGCCAUUUACACUCCCAAGCUCCCAACCAGCCGCCUUAUCCUU---UCUCUCUCUGUGCUUUC------------- .(((((...(((..((((.(((((.((..((((((......))))))......((...............)).....))))).)).))))..)))---...))).))........------------- ( -23.16, z-score = -0.67, R) >droEre2.scaffold_4820 6370471 105 + 10470090 AAGCAGACUGAGUCUGAGGCGCAGGAUCCACCGCAUAUCCUUGCGGUACCCCCGCCAAUUCCACU-------UCCCAACCACCCGCUUUAUCCUU---UCUUCCUCUGCACUUUC------------- ..(((((..(((..(((((((..(....)((((((......))))))..................-------...........)))))))..)))---......)))))......------------- ( -25.40, z-score = -2.85, R) >dp4.chr2 22656006 121 + 30794189 ------AACAAGCAUAACGCGCGCAGGCCACCACAUAUCCUUGCGGUUCUCUCUCUCUCUUUCCUUCAU-UUUCGCACACUCACACACUGUCUUUGUACUUUCCUUUGUGGCCCUGCGUGUGUGUGUG ------.....(((((.((((((((((((((...........((((.......................-..))))(((...((.....))...)))..........)))).))))))))))))))). ( -32.81, z-score = -3.55, R) >droPer1.super_0 9849507 109 + 11822988 ------AACAAGCAUGACGCGCGCAGGCCACCACAUAUCCUUGCGGUUCUCUCUCUCUCU--CCUUCUU-UUUCGCACACUCACACAGUGUCUUUGUACUUUCCUUUGCGGCCCUGUG---------- ------.....((.....)).(((((((((((.((......)).))).............--.......-....(.(((((.....)))))).................)).))))))---------- ( -19.90, z-score = -0.45, R) >consensus AAG_AGACUGAGCCUGAGGCGCAGAGUCCACCGCAUAUCCUUGCGGUUCUCCCGCCAUUUCCACUUCUCACCUCCCAUCCACCCUCCUUAUCCUU___UCUCUCUCUGUACUUUC_____________ ....................((((((...((((((......))))))......(.......).........................................))))))................... ( -7.28 = -8.26 + 0.98)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:03 2011