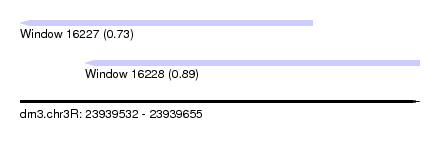
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,939,532 – 23,939,655 |
| Length | 123 |
| Max. P | 0.887006 |

| Location | 23,939,532 – 23,939,622 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.19 |
| Shannon entropy | 0.60780 |
| G+C content | 0.39326 |
| Mean single sequence MFE | -15.75 |
| Consensus MFE | -10.07 |
| Energy contribution | -9.71 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.47 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23939532 90 - 27905053 CGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCUACAAAAACCG---GCAGCCG------CUUUGGUCCACCCCA------------ .(((((((.....))))))).....((((...(((((((........)))))))....))))..((((---((....)------))..)))........------------ ( -14.70, z-score = -0.40, R) >droSim1.chr3R 23639243 90 - 27517382 CGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCUACAAAAACCG---GCAGCCG------CUUUGGUCCACCCCA------------ .(((((((.....))))))).....((((...(((((((........)))))))....))))..((((---((....)------))..)))........------------ ( -14.70, z-score = -0.40, R) >droSec1.super_22 558243 89 - 1066717 CGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCUACAAAAACCG---GCAGCU-------CUUUGGUCCACCCCA------------ .(((((((.....))))))).....((((...(((((((........)))))))....))))..((((---(.....-------..)))))........------------ ( -12.80, z-score = -0.29, R) >droYak2.chr3R 6275734 90 + 28832112 CUAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCUACAAAAACCG---GCAGCCG------CUUUGGUCCACCCCA------------ ..((((((.....))))))......((((...(((((((........)))))))....))))..((((---((....)------))..)))........------------ ( -10.60, z-score = 0.90, R) >droEre2.scaffold_4820 6366190 89 + 10470090 CGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCUAC-AAAACCG---GCAGCCG------CUUUGGUCCACCCCA------------ .(((((((.....))))))).....((((...(((((((........)))))))....))-)).((((---((....)------))..)))........------------ ( -14.70, z-score = -0.45, R) >droAna3.scaffold_13340 22081691 83 - 23697760 AGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCUACGACAG-------CAGUGGCCCAUCCCCCCA--------------------- ((((((((.....))))))))...........(((((((........)))))))..((((.....-------..))))............--------------------- ( -12.50, z-score = -0.65, R) >dp4.chr2 22651169 108 + 30794189 UGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCUCGGUAAUAAGCAACCUACGAAAGCAG--CACAGUAGCACAGCAGCAGAGCCGCCGCCGGCAUCACUCU- .(((((((.....)))))))....(((((...(((((((........)))))))..((.(....).))--.))))).((......)).((((((....)))).)).....- ( -20.70, z-score = 0.28, R) >droPer1.super_0 9844645 100 + 11822988 UGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCUCGGUAAUAAGCAACCUACGAAAGCAG--CACAGU--------AGCAGAGCCGCCGCCGGCACCACUCU- .(((((((.....)))))))......(((((.(((...((((........))))....))).)))))(--(.....--------.))...((((....))))........- ( -19.00, z-score = -0.16, R) >droWil1.scaffold_181108 1174571 93 - 4707319 AGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCAGCAGCGGGCACCAUC--CCAGC-AUCAGC--ACCCACCAU------------- ((((((((.....))))))))...........((((.((((..(((........)))......(((......)--)).))-)).)))--)........------------- ( -19.10, z-score = -2.12, R) >droVir3.scaffold_13047 3618646 104 + 19223366 AGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCGGCAGCAG---CCAUU--UCAUCGAGCCAC--GCCCACCAUCAUCUCCCUACCC ((((((((.....))))))))......((((.(((((((........)))))))...(((....)---))...--.....))))...--...................... ( -15.90, z-score = -1.20, R) >droMoj3.scaffold_6263 18760 105 - 24457 CAAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGUACGGUAAUAAGCAACCGGCAGCAGUUAUAACCAACGGCCCAUCGCCGCUCUCACUCUGCCGCCU------ ..((((((.....)))))).............(((((((........)))))))...(((.((((..........((((.....))))........)))).))).------ ( -21.77, z-score = -1.74, R) >droGri2.scaffold_14830 4385347 99 - 6267026 CGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCAGCAGCCG-CACCACCAACCAGCCAGCCAUUGAGCCACCAGCC----------- .(((((((.....))))))).......((((((((((((........)))))))....((....)-)....................)))))........----------- ( -12.50, z-score = 0.54, R) >consensus CGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCUACAAAAGCCG___GCAGCCG______CUCUGGUCCACCCCA____________ .(((((((.....))))))).....((((...(((((((........)))))))....))))................................................. (-10.07 = -9.71 + -0.36)

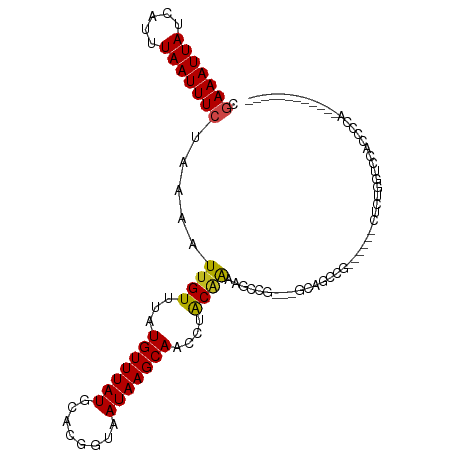
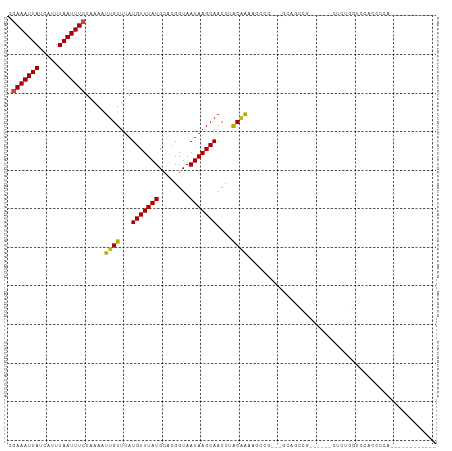
| Location | 23,939,552 – 23,939,655 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 85.29 |
| Shannon entropy | 0.31335 |
| G+C content | 0.29492 |
| Mean single sequence MFE | -21.03 |
| Consensus MFE | -14.93 |
| Energy contribution | -15.18 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23939552 103 - 27905053 ------GCCGAGAUAAUGCAUAAAAUUAUGUAAAUAUUCCGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCUACAAAAACCGGC------ ------((((......((((((((.....(((((((....(((((((.....)))))))......))))))).)))))))).(((........))).........))))------ ( -22.00, z-score = -2.82, R) >droSim1.chr3R 23639263 103 - 27517382 ------GUCGAGAUAAUGCAUAAAAUUAUGUAAAUAUUCCGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCUACAAAAACCGGC------ ------((((......((((((((.....(((((((....(((((((.....)))))))......))))))).)))))))).(((........))).........))))------ ( -19.30, z-score = -2.04, R) >droSec1.super_22 558262 103 - 1066717 ------GUCAAGAUAAUGCAUAAAAUUAUGUAAAUAUUCCGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCUACAAAAACCGGC------ ------(((.......((((((((.....(((((((....(((((((.....)))))))......))))))).)))))))).(((........)))..........)))------ ( -17.50, z-score = -1.66, R) >droYak2.chr3R 6275754 103 + 28832112 ------GCCGAGAUAAUGCAUAAAAUUAUGUAAAUAUUCCUAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCUACAAAAACCGGC------ ------((((......((((((((.....(((((((.....((((((.....)))))).......))))))).)))))))).(((........))).........))))------ ( -18.70, z-score = -1.85, R) >droEre2.scaffold_4820 6366210 102 + 10470090 ------GCCGAGAUAAUGCAUAAAAUUAUGUAAAUAUUCCGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCUAC-AAAACCGGC------ ------((((......((((((((.....(((((((....(((((((.....)))))))......))))))).)))))))).(((........)))...-.....))))------ ( -22.00, z-score = -2.80, R) >droAna3.scaffold_13340 22081708 101 - 23697760 ----CCGUCGAGAUAAUGCAUAAAAUUAUGUAAAUAUUCAGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCUACGACAGC---------- ----..((((......((((((((.....(((((((...((((((((.....)))))))).....))))))).)))))))).(((........)))..))))...---------- ( -23.70, z-score = -3.61, R) >dp4.chr2 22651203 113 + 30794189 CCGUCGAGAGAGAUAAUGCAUAAAAUUAUGUAAAUAUUCUGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCUCGGUAAUAAGCAACCUACGAAAGCAGCACAGU-- (((((....))......(((((((.....(((((((....(((((((.....)))))))......))))))).))))))).)))...........((.(....).))......-- ( -18.90, z-score = -0.55, R) >droPer1.super_0 9844671 113 + 11822988 CCGUCGAGAGAGAUAAUGCAUAAAAUUAUGUAAAUAUUCUGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCUCGGUAAUAAGCAACCUACGAAAGCAGCACAGU-- (((((....))......(((((((.....(((((((....(((((((.....)))))))......))))))).))))))).)))...........((.(....).))......-- ( -18.90, z-score = -0.55, R) >droWil1.scaffold_181108 1174594 105 - 4707319 ----CCGUCGGGAUAAUGCAUAAAAUUAUGUAAAUAUUCAGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCAGCAGCGGGCACC------ ----((((.(......((((((((.....(((((((...((((((((.....)))))))).....))))))).)))))))).(((........)))..).)))).....------ ( -23.50, z-score = -1.99, R) >droVir3.scaffold_13047 3618675 109 + 19223366 -----GUUGGGGAUAAUGCAUAAAAUUAUGUAAAUAUUCAGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCGGCAGCAGCCAUUUCAUC- -----....(..((..((((((((.....(((((((...((((((((.....)))))))).....))))))).)))))))).(((........)))(((....)))))..)...- ( -24.60, z-score = -2.41, R) >droMoj3.scaffold_6263 18789 110 - 24457 -----GUUGGGGAUAAUGCAUAAAAUUAUGUAAAUAUUCCAAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGUACGGUAAUAAGCAACCGGCAGCAGUUAUAACCAAC -----(((((..(((((((.......(((((((((((..((((((((.....))))).......)))...)))))))))))((((........))))...))).))))..))))) ( -22.81, z-score = -1.80, R) >droGri2.scaffold_14830 4385371 109 - 6267026 -----UUUAGGGAUAAUGCAUAAAAUUAUGUAAAUAUUCCGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCAGCAGCCG-CACCACCAAC -----....((((((.((((((....))))))..))))))(((((((.....))))))).....(((((..(((((((........)))))))...)))))...-.......... ( -20.40, z-score = -1.44, R) >consensus ______GUCGAGAUAAUGCAUAAAAUUAUGUAAAUAUUCCGAAAUUAUCAUUUAAUUUCUAAAAUUGUUUAUGUUUAUGCACGGUAAUAAGCAACCUACAAAAGCCGGC______ ................((((((((.....(((((((....(((((((.....)))))))......))))))).)))))))).(((........)))................... (-14.93 = -15.18 + 0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:02 2011