| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,923,113 – 23,923,203 |
| Length | 90 |
| Max. P | 0.557096 |

| Location | 23,923,113 – 23,923,203 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 78.28 |
| Shannon entropy | 0.46684 |
| G+C content | 0.36303 |
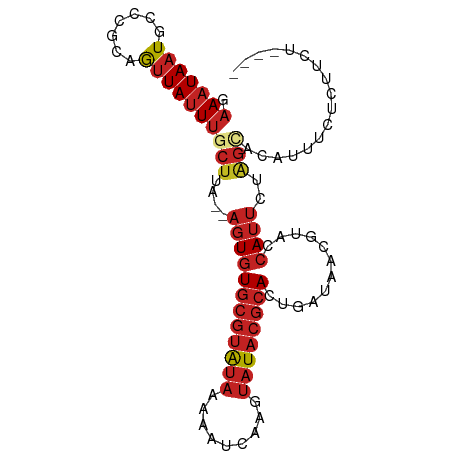
| Mean single sequence MFE | -17.01 |
| Consensus MFE | -7.77 |
| Energy contribution | -7.96 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.557096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
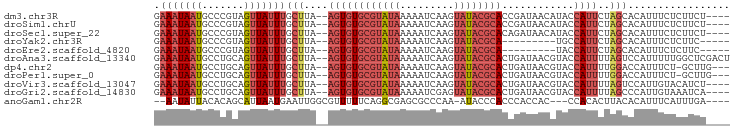
>dm3.chr3R 23923113 90 - 27905053 GAAAUAAUGCCCGUAGUUAUUUGCUUA--AGUGUGCGUAUAAAAAUCAAGUAUACGCACCGAUAACAUACCAUUCUAGCACAUUUCUCUUCU---- (((((..(((..((((....))))...--.(.(((((((((.........)))))))))).................))).)))))......---- ( -17.00, z-score = -2.31, R) >droSim1.chrU 14881017 90 + 15797150 GAAAUAAUGCCCGUAGUUAUUUGCUUA--AGUGUGCGUAUAAAAAUCAAGUAUACGCACCGAUAACAUACCAUUCUAGCACAUUUCUCUUCU---- (((((..(((..((((....))))...--.(.(((((((((.........)))))))))).................))).)))))......---- ( -17.00, z-score = -2.31, R) >droSec1.super_22 541879 90 - 1066717 GAAAUAAUGCCCGUAGUUAUUUGCUUA--AGUGUGCGUAUAAAAAUCAAGUAUACGCACAGAUAACAUACCAUUCUAGCACAUUUCUCUUCU---- (((((..(((..((((((((((((...--.))(((((((((.........)))))))))))))))).))).......))).)))))......---- ( -19.00, z-score = -2.91, R) >droYak2.chr3R 6258623 80 + 28832112 GAAAUAAUGCCCGUAGUUAUUUGCUUA--AGUGUGCGUAUAAAAAUCAAGUAUACGCA---------UGCCAUUCUAGCACAUUUCUCUUC----- (((((..(((..((((....))))...--.(((((((((((.........))))))))---------))).......))).))))).....----- ( -18.00, z-score = -2.85, R) >droEre2.scaffold_4820 6347771 80 + 10470090 GAAAUAAUGCCCGUAGUUAUUUGCUUA--AGUGUGCGUAUAAAAAUCAAGUAUACGCA---------UACCAUUCUAGCACAUUUCUCUUC----- (((((..(((..((((....))))...--.(((((((((((.........))))))))---------))).......))).))))).....----- ( -19.00, z-score = -3.84, R) >droAna3.scaffold_13340 22065070 94 - 23697760 GAAAUAAUGCCUGCAGUUAUUUGCUUA--AGUGUGCGUAUAAAAAUCAAGUAUACGCACUGAUAACGUACCAUUUUAGUCCAUUUUUGGCUCGACU ........(((.((((....))))...--...(((((((((.........)))))))))............................)))...... ( -18.40, z-score = -0.65, R) >dp4.chr2 22634332 90 + 30794189 GAAAUAAUGCCUGCAGUUAUUUGCUUA--AGUGUGCGUAUAAAAAUCAAGUAUACGCACUGAUAACGUACCAUUUUGGACCAUUUCU-GCUUG--- ............((((..((.((((((--...(((((((((.........)))))))))...))).)))((.....))...))..))-))...--- ( -18.10, z-score = -0.91, R) >droPer1.super_0 9827690 90 + 11822988 GAAAUAAUGCCUGCAGUUAUUUGCUUA--AGUGUGCGUAUAAAAAUCAAGUAUACGCACUGAUAACGUACCAUUUUGGACCAUUUCU-GCUUG--- ............((((..((.((((((--...(((((((((.........)))))))))...))).)))((.....))...))..))-))...--- ( -18.10, z-score = -0.91, R) >droVir3.scaffold_13047 3580865 90 + 19223366 GAAAUAAUGCCUGCAGUUAUUUGCUUA--AGUGUGCGUAUAAAAAUCAAGUAUACGCACUGAUAACGUACCAUUUUAGUCCAUUGUACAUCU---- ...((((((...((((....))))...--...(((((((((.........))))))))).....................))))))......---- ( -17.00, z-score = -0.96, R) >droGri2.scaffold_14830 4364627 90 - 6267026 GAAAUAAUGCCUGCAGUUAUUUGCUUA--AGUGUGCGUAUAAAAAUCGAGUAUACGCACUGAUAACGUACCAUUUUAGCCCAUUGUAAAUCA---- ...((((((...((((....))))...--...(((((((((.........))))))))).....................))))))......---- ( -17.00, z-score = -0.88, R) >anoGam1.chr2R 19462569 86 + 62725911 --AAUAUUACACAGCAUUAAUGAAUUGGCGUUUUUCAGGCGAGCGCCCAA-AUACCCACCCACCAC---CCACACUUACACAUUUCAUUUGA---- --........................(((((((.......)))))))...-...............---.......................---- ( -8.50, z-score = 0.20, R) >consensus GAAAUAAUGCCCGCAGUUAUUUGCUUA__AGUGUGCGUAUAAAAAUCAAGUAUACGCACUGAUAACGUACCAUUCUAGCACAUUUCUCUUCU____ ............((((....)))).........((((((((.........))))))))...................................... ( -7.77 = -7.96 + 0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:59 2011