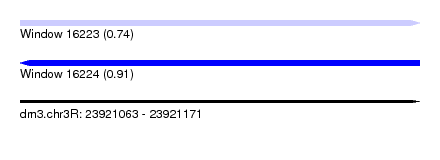
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,921,063 – 23,921,171 |
| Length | 108 |
| Max. P | 0.914945 |

| Location | 23,921,063 – 23,921,171 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.98 |
| Shannon entropy | 0.35566 |
| G+C content | 0.48335 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -16.38 |
| Energy contribution | -17.22 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.740534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
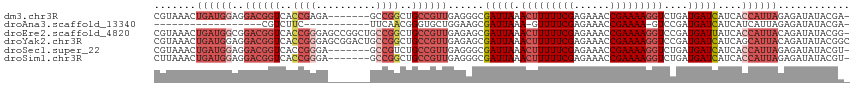
>dm3.chr3R 23921063 108 + 27905053 -UCGUAUAUCUCUAAUGGUGAUGAUCAUCAGACCUUUUCGGUUUCUCGAAAAAGUUUAAUCGCCCUCAACGGCAGCCGGC-------UCUCGGUGACCGUCCUCCAUCAGUUUACG -.((((.....((.((((.((((.(((((((((((((((((....))))))).........(((......)))....)).-------))).))))).))))..)))).))..)))) ( -29.90, z-score = -2.47, R) >droAna3.scaffold_13340 22062784 84 + 23697760 -UCGUAUAUCUCUAAUGAUGAUGAUCAUCGGAC-UUUUCGGUUUCUCGAAAAC-UUUAAUCGCUUCCAGCACCCGUUGAA-----------GAAGACG------------------ -((((.((((......))))))))(((.(((..-(((((((....))))))).-.......((.....))..))).))).-----------.......------------------ ( -15.80, z-score = 0.07, R) >droEre2.scaffold_4820 6345672 115 - 10470090 -CCGUAUAUCUGUAAUGGUGAUAAUCAUCGGACCUUUUCGGUUUCUCGAAAAAGUUUAAUCGCUCUCAACGGCAGCCGGCAGCCGGCUCCCGGUGACCGUCCGCCAUCAGUUUACG -.((((...(((..((((((((..(((((((((.(((((((....))))))).))......(((......)))((((((...)))))).)))))))..)).)))))))))..)))) ( -38.60, z-score = -3.10, R) >droYak2.chr3R 6256490 116 - 28832112 GCCGUAUAUCUGUAAUGCUGAUGAUCAUCGGACCUUUUCGGUUUCUCGAAAAAGUUUAAUCGCUCUCAACGGCAGCCGGCAGUCCGCUCCCGGUGACCGUCCUCCAUCAGUUUACG ...........((((.(((((((...((..(((.(((((((....))))))).)))..))........((((..(((((.((....)).)))))..))))....))))))))))). ( -35.70, z-score = -2.90, R) >droSec1.super_22 539826 108 + 1066717 -ACGUAUAUCUCUAAUGGUGAUGAUCAUCAGACCUUUUCGGUUUCUCGAAAAAGUUUAAUCGCCCUCAACGGCAGACGGC-------UCCCGGUGACCGUCCUCCAUCAGUUUACG -.((((.....((.((((.((((.(((((((((.(((((((....))))))).))))....(((.((.......)).)))-------....))))).))))..)))).))..)))) ( -30.30, z-score = -2.30, R) >droSim1.chr3R 23621191 108 + 27517382 -ACGUAUAUCUCUAAUGGUGAUGAUCAUCAGACCUUUUCGGUUUCUCGAAAAAGUUUAAUCGCCCUCAACGGCAGCCGGC-------UCCCGGUGACCGUCCUCCAUCAGUUUAAG -..........((.((((.((((.((...((((.(((((((....))))))).))))....(((......))).(((((.-------..))))))).))))..)))).))...... ( -27.30, z-score = -1.66, R) >consensus _ACGUAUAUCUCUAAUGGUGAUGAUCAUCAGACCUUUUCGGUUUCUCGAAAAAGUUUAAUCGCCCUCAACGGCAGCCGGC_______UCCCGGUGACCGUCCUCCAUCAGUUUACG ................((((.....))))((((.(((((((....))))))).))))...........((((..(((((..........)))))..))))................ (-16.38 = -17.22 + 0.84)

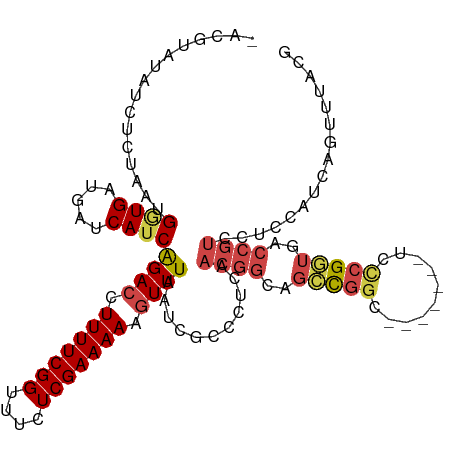
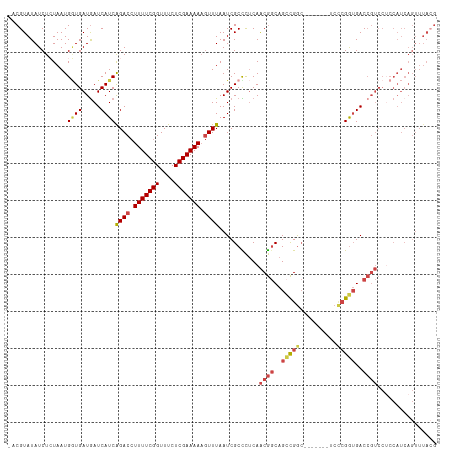
| Location | 23,921,063 – 23,921,171 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.98 |
| Shannon entropy | 0.35566 |
| G+C content | 0.48335 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -19.69 |
| Energy contribution | -22.50 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23921063 108 - 27905053 CGUAAACUGAUGGAGGACGGUCACCGAGA-------GCCGGCUGCCGUUGAGGGCGAUUAAACUUUUUCGAGAAACCGAAAAGGUCUGAUGAUCAUCACCAUUAGAGAUAUACGA- ((((..(((((((..((((((..(((...-------..)))..))))))((.((..((((.(((((((((......))))))))).))))..)).)).))))))).....)))).- ( -35.60, z-score = -3.03, R) >droAna3.scaffold_13340 22062784 84 - 23697760 ------------------CGUCUUC-----------UUCAACGGGUGCUGGAAGCGAUUAAA-GUUUUCGAGAAACCGAAAA-GUCCGAUGAUCAUCAUCAUUAGAGAUAUACGA- ------------------(((..((-----------((...(((...((((((((.......-)))))).))...)))....-....((((.....))))....))))...))).- ( -18.90, z-score = -0.27, R) >droEre2.scaffold_4820 6345672 115 + 10470090 CGUAAACUGAUGGCGGACGGUCACCGGGAGCCGGCUGCCGGCUGCCGUUGAGAGCGAUUAAACUUUUUCGAGAAACCGAAAAGGUCCGAUGAUUAUCACCAUUACAGAUAUACGG- ((((..((((((((((((..(((.(((.((((((...)))))).))).)))............(((((((......)))))))))))).((.....))))))..)))...)))).- ( -39.50, z-score = -2.57, R) >droYak2.chr3R 6256490 116 + 28832112 CGUAAACUGAUGGAGGACGGUCACCGGGAGCGGACUGCCGGCUGCCGUUGAGAGCGAUUAAACUUUUUCGAGAAACCGAAAAGGUCCGAUGAUCAUCAGCAUUACAGAUAUACGGC .((((.(((((((..((((((..((((.((....)).))))..)))))).......(((..(((((((((......)))))))))..)))..)))))))..))))........... ( -38.30, z-score = -2.36, R) >droSec1.super_22 539826 108 - 1066717 CGUAAACUGAUGGAGGACGGUCACCGGGA-------GCCGUCUGCCGUUGAGGGCGAUUAAACUUUUUCGAGAAACCGAAAAGGUCUGAUGAUCAUCACCAUUAGAGAUAUACGU- ((((..(((((((..((.(((((.(((..-------(((.((.......)).)))......(((((((((......)))))))))))).))))).)).))))))).....)))).- ( -36.30, z-score = -2.65, R) >droSim1.chr3R 23621191 108 - 27517382 CUUAAACUGAUGGAGGACGGUCACCGGGA-------GCCGGCUGCCGUUGAGGGCGAUUAAACUUUUUCGAGAAACCGAAAAGGUCUGAUGAUCAUCACCAUUAGAGAUAUACGU- ......(((((((..((((((..((((..-------.))))..))))))((.((..((((.(((((((((......))))))))).))))..)).)).)))))))..........- ( -36.30, z-score = -2.93, R) >consensus CGUAAACUGAUGGAGGACGGUCACCGGGA_______GCCGGCUGCCGUUGAGAGCGAUUAAACUUUUUCGAGAAACCGAAAAGGUCCGAUGAUCAUCACCAUUAGAGAUAUACGA_ .......((((((..((((((..((((..........))))..))))))......(((((.(((((((((......)))))))))....)))))....))))))............ (-19.69 = -22.50 + 2.81)
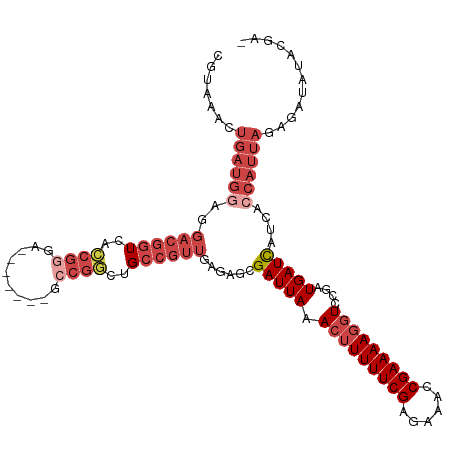
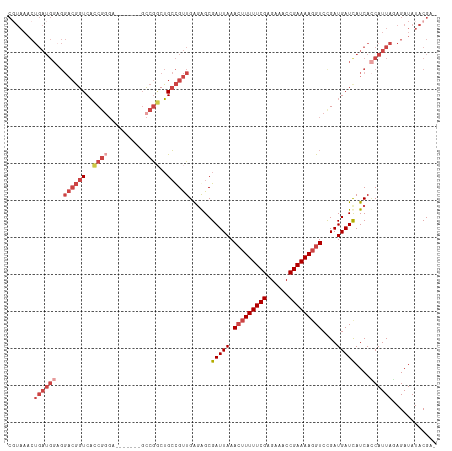
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:58 2011