| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,920,940 – 23,921,045 |
| Length | 105 |
| Max. P | 0.749438 |

| Location | 23,920,940 – 23,921,045 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.83 |
| Shannon entropy | 0.43483 |
| G+C content | 0.50695 |
| Mean single sequence MFE | -34.29 |
| Consensus MFE | -24.06 |
| Energy contribution | -23.64 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
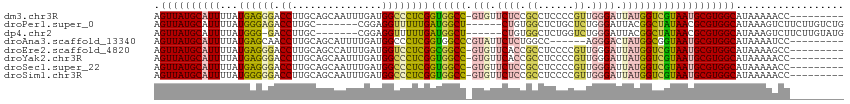
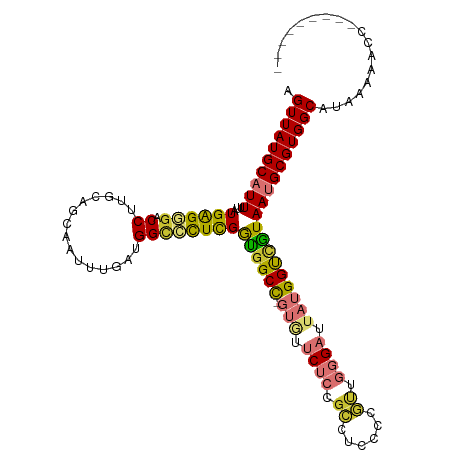
>dm3.chr3R 23920940 105 + 27905053 AGUUAUGCAUUUUAUGAGGGACCUUGCAGCAAUUUGAUGGCCCUCGGUGGCC-GUGUUCUCCGCCUCCCCGUUGGGAUUAUGGUCGUAAUGCGUGGCAUAAAAACC--------- .((((((((((...((((((.((...(((....)))..))))))))..((((-(((.((((.((......)).)))).)))))))..)))))))))).........--------- ( -38.20, z-score = -2.11, R) >droPer1.super_0 9825795 102 - 11822988 AGUUAUGCAUUUUAUGGGAGACCUUGC-------CGGAGGUUUUUGAUGGCU------CUGUGGCUCUGCUCUGGGAUUACGGCUAUAACGCGUGGCAUAAAGUCUUCUUGUCUG .(((((((........(((((((((..-------..))))))))).((((((------..(((((((......))).))))))))))...))))))).................. ( -27.00, z-score = 0.03, R) >dp4.chr2 22632464 101 - 30794189 AGUUAUGCAUUUUAUGGG-GACCUUGC-------CGGAGGUUUUUGAUGGCU------CUGUGGCUCUGGUCUGGGAUUACGGCUAUAACGCGUGGCAUAAAGUCUUCUUGUAUG .(((((((...(((((((-((((..((-------((.(((((......))).------)).))))...)))))(......)..)))))).))))))).................. ( -28.90, z-score = -0.88, R) >droAna3.scaffold_13340 22062652 100 + 23697760 AGUUAUGCAUUUUAUGAGCAACCUUGCAGCAUUUUGAUGGCCCUCGGUGGCCCGUAUUCUCUGGCC------AGGGACUAUGGCGGUAAUGCGUGGCAUAAAAUCC--------- .((((((((((......(((....))).((........((((......))))((((.(((((....------))))).))))))...)))))))))).........--------- ( -29.80, z-score = 0.18, R) >droEre2.scaffold_4820 6345549 105 - 10470090 AGUUAUGCAUUUUAUGAGGGACCUUGCAGCCAUUUGAUGGUCCUCGGCGGCC-GUGUUCACCGCCUCCCCGUUGGGAUUAUGGUCGUAAUGCGUGGCAUAAAAGCC--------- .((((((((((...(((((.(((...(((....)))..))))))))((((((-(((.((.(((.(.....).))))).))))))))))))))))))).........--------- ( -38.30, z-score = -1.60, R) >droYak2.chr3R 6256367 105 - 28832112 AGUUAUGCAUUUUAUGAGGGACCUUGCAGCAAUUUGAUGGCCCUCGGUGGCC-GUGUUCACCGCCUCCCCGUUGGGAUUAUGGUCGUAAUGCGUGGCAUAAAAACC--------- .((((((((((...((((((.((...(((....)))..))))))))..((((-(((.((.(((.(.....).))))).)))))))..)))))))))).........--------- ( -36.60, z-score = -1.47, R) >droSec1.super_22 539703 105 + 1066717 AGUUAUGCAUUUUAUGAGGGACCUUGCAGCAAUUUGAUGGCCCUCGGUGGCC-GUGUUCUCCGCCUCCCCGUUGGGAUUAUGGUCGUAAUGCGUGGCAUAAAAACC--------- .((((((((((...((((((.((...(((....)))..))))))))..((((-(((.((((.((......)).)))).)))))))..)))))))))).........--------- ( -38.20, z-score = -2.11, R) >droSim1.chr3R 23621068 105 + 27517382 AGUUAUGCAUUUUAUGGGGGACCUUGCAGCAAUUUGAUGGCCCUCGGUGGCC-GUGUUCUCCGCCUCCCCGUUGGGAUUAUGGUCGUAAUGCGUGGCAUAAAAACC--------- .((((((((((...((((((.((...(((....)))..))))))))..((((-(((.((((.((......)).)))).)))))))..)))))))))).........--------- ( -37.30, z-score = -1.21, R) >consensus AGUUAUGCAUUUUAUGAGGGACCUUGCAGCAAUUUGAUGGCCCUCGGUGGCC_GUGUUCUCCGCCUCCCCGUUGGGAUUAUGGUCGUAAUGCGUGGCAUAAAAACC_________ .((((((((((...((((((.((...(((....)))..))))))))((((((.....((((.((......)).))))....)))))))))))))))).................. (-24.06 = -23.64 + -0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:57 2011