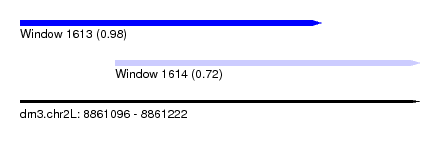
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,861,096 – 8,861,222 |
| Length | 126 |
| Max. P | 0.982522 |

| Location | 8,861,096 – 8,861,191 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.95 |
| Shannon entropy | 0.40301 |
| G+C content | 0.48003 |
| Mean single sequence MFE | -22.22 |
| Consensus MFE | -18.27 |
| Energy contribution | -18.13 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.982522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
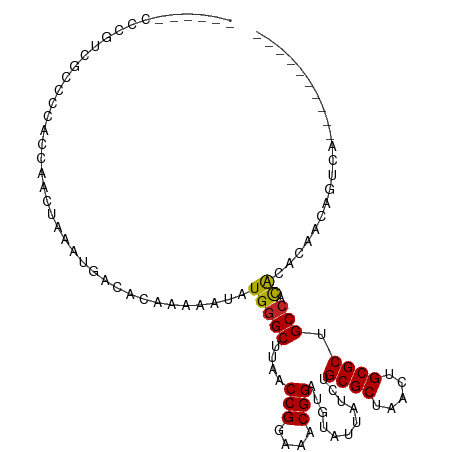
>dm3.chr2L 8861096 95 + 23011544 ------CCCGUCGCCCCACCAACUAAAUGACACAAAAAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCACACACAACAGUCA---------- ------.....................((((.((......))(((....(((....)))...........((((.....)))).)))..........))))---------- ( -21.50, z-score = -1.34, R) >droSim1.chr2L 8640055 95 + 22036055 ------CCCGUCGGCCCACCAACUAAAUGACACAAAAAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCACACACAACAGUCA---------- ------...((.((....)).))....((((.((......))(((....(((....)))...........((((.....)))).)))..........))))---------- ( -22.90, z-score = -1.36, R) >droSec1.super_3 4335199 95 + 7220098 ------CCCGUCGGCCCACCAACUAAAUGACACAAAAAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCACACACAACAGUCA---------- ------...((.((....)).))....((((.((......))(((....(((....)))...........((((.....)))).)))..........))))---------- ( -22.90, z-score = -1.36, R) >droYak2.chr2L 11508063 99 + 22324452 ------CCCGUCGCCCCACCAACUAAAUGACACAAAAAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCACACACAACAGUCAGUCG------ ------.....................((((.((......))(((....(((....)))...........((((.....)))).)))..........))))....------ ( -22.30, z-score = -1.11, R) >droEre2.scaffold_4929 9454263 95 + 26641161 ------CCCUUCGCCCCACCGACUGAAUGACACAAAAAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCACACACAACAGUCA---------- ------..............(((((.......((......))(((....(((....)))...........((((.....)))).)))........))))).---------- ( -25.50, z-score = -2.93, R) >droAna3.scaffold_12916 10525058 99 + 16180835 ------CCCCCUCUCCAGUAAACUAAAUGACACAAAAAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCACACACAACAGUCAGUCA------ ------.....................((((.((......))(((....(((....)))...........((((.....)))).)))..........))))....------ ( -22.30, z-score = -1.66, R) >dp4.chr4_group5 1660627 109 - 2436548 --CCUAUCCCCACAUACACACACUAAAUGACACAAAAAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCACACACAACAUUCAGUCAGCCAGU --...................(((...((((.((......))(((....(((....)))...........((((.....)))).)))..............))))...))) ( -23.50, z-score = -1.83, R) >droPer1.super_5 1631994 103 - 6813705 --CCUAUCCCCACAUACAUACACUAAAUGACACAAAAAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCACACACAACAUUCAGUCA------ --.........................((((.((......))(((....(((....)))...........((((.....)))).)))..............))))------ ( -21.50, z-score = -2.29, R) >droWil1.scaffold_180772 8718800 84 - 8906247 ------CACUGCUGGAUGCUCAGUAAAUGACACAAAAAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCACA--------------------- ------...((((((....)))))).................(((....(((....)))...........((((.....)))).)))...--------------------- ( -23.40, z-score = -2.33, R) >droMoj3.scaffold_6500 10861610 90 + 32352404 CUGUCCCUUUACUCUCUACUUACUAAAUGACACAAAAAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCUCG--------------------- .((((..((((............)))).)))).........((((....(((....)))...........((((.....)))).))))..--------------------- ( -19.50, z-score = -3.26, R) >droGri2.scaffold_15126 7879586 89 + 8399593 ------CUCUUCCCUGCCCCACCUAAAUGACACAAAAAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCGCUGCCUCG---------------- ------...................................((((....(((....)))...........((((.....))))......))))..---------------- ( -19.10, z-score = -1.37, R) >consensus ______CCCGUCGCCCCACCAACUAAAUGACACAAAAAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCACACACAACAGUCA__________ ........................................(((((....(((....)))...........((((.....)))).))).))..................... (-18.27 = -18.13 + -0.14)

| Location | 8,861,126 – 8,861,222 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.19 |
| Shannon entropy | 0.37006 |
| G+C content | 0.54722 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -20.02 |
| Energy contribution | -21.18 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.724904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
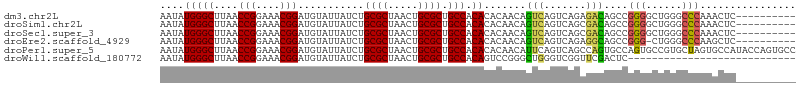
>dm3.chr2L 8861126 96 + 23011544 AAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCACACACAACAGUCAGUCAGAGACAGCCGGGGCUGGGCCCAAACUC---------- ....((((((...(((....)))........((((.(((.((((...((.....))...)))).)))))))..((((....)))))))))).....---------- ( -32.70, z-score = -1.98, R) >droSim1.chr2L 8640085 96 + 22036055 AAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCACACACAACAGUCAGUCAGCGACAGCCGGGGCUGGGCCCAAACUC---------- ....((((((...(((....)))............((((.((((.((((..........)))))))).)))).((((....)))))))))).....---------- ( -36.50, z-score = -2.96, R) >droSec1.super_3 4335229 96 + 7220098 AAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCACACACAACAGUCAGUCAGCGACAGCCGGGGCUGGGCCCAAACUC---------- ....((((((...(((....)))............((((.((((.((((..........)))))))).)))).((((....)))))))))).....---------- ( -36.50, z-score = -2.96, R) >droEre2.scaffold_4929 9454293 95 + 26641161 AAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCACACACAACAGUCAGUCAGAGGCAGCCGGG-CUGGGCCCAAGCUC---------- ....((((((((.(((....)))..................(((.((((((.(..((........))..).))))))))).-.)))))))).....---------- ( -33.40, z-score = -1.38, R) >droPer1.super_5 1632028 106 - 6813705 AAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCACACACAACAUUCAGUCAGCCAGUGCCAGUGCCGUGCUAGUGCCAUACCAGUGCC ......(((....(((....)))...........((((((((.((((((.(((.....((.....))......))).)))))).))..)))))).........))) ( -29.60, z-score = 0.49, R) >droWil1.scaffold_180772 8718830 78 - 8906247 AAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCACAGUCCGGGCUGGGUCGGUUCGACUC---------------------------- ......(((....(((....)))...........((((.....)))).))).((((....))))(((((...))))).---------------------------- ( -26.90, z-score = -1.67, R) >consensus AAUAUGGGCUUAACCGGAAACGGAUGUAUUAUCUGCGCUAACUGCGCUGCCACACACAACAGUCAGUCAGCGACAGCCGGGGCUGGGCCCAAACUC__________ ....(((((....(((....)))...........((((.....)))).))).))...................((((....))))..................... (-20.02 = -21.18 + 1.17)
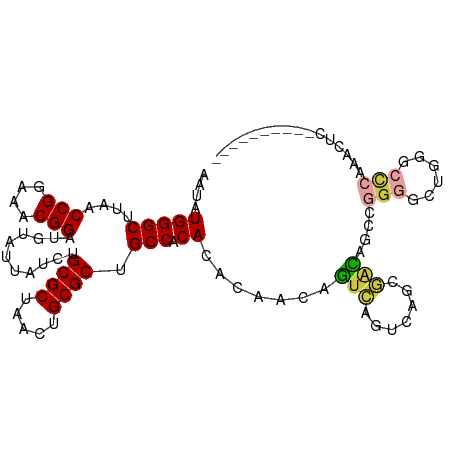
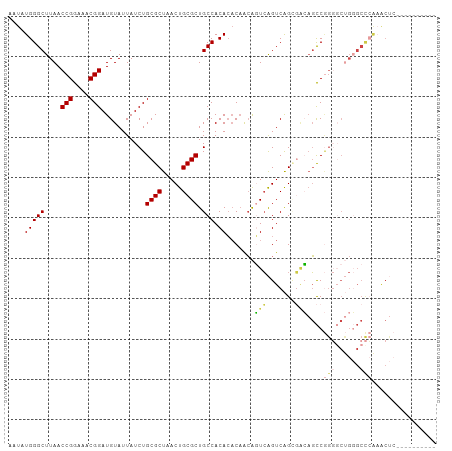
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:07 2011