
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,909,113 – 23,909,212 |
| Length | 99 |
| Max. P | 0.589985 |

| Location | 23,909,113 – 23,909,212 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 78.12 |
| Shannon entropy | 0.41976 |
| G+C content | 0.67240 |
| Mean single sequence MFE | -48.53 |
| Consensus MFE | -25.82 |
| Energy contribution | -26.21 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23909113 99 - 27905053 GGGUAGGUGGGCGUGGGCGAGUACCAGCAGCCCGUGGCGGCCAGCUGCUGUCCGUAGGCGGCGGCGGC---AGCAGCCGCGCCCACUCCAUACGGGAAACCG .((..(((((((..((((..((....)).))))(((((.(((.(((((((((....))))))))))))---....))))))))))))))...(((....))) ( -59.60, z-score = -3.00, R) >droAna3.scaffold_13340 22050287 99 - 23697760 GGGUAGGUGGGCGUCGGCGAGUACCAGCAUCCCGUGGCGGCUAGCUGUUGACCGUAGGCGGCGGCAGC---AGCGGCGGCGCCCACUCCAUACGGGAAUCCU ((((..(((((((((((.((((....)).))))...((.(((.(((((((.(((....))))))))))---))).)))))))))))(((.....))))))). ( -51.60, z-score = -1.86, R) >dp4.chr2 22618810 96 + 30794189 UGGUAUGUGGGUGUGGGUGAGUACCAGCAGCCGGUGGCCGCCAGCUGCUGGCCGUAAGCGGCGGC------AGCAGUGGCCCCUAAGCCGUAUGGGAACCCG .(((....((.(((.(((....))).))).))((.((((((..(((((((.(((....)))))))------))).))))))))...)))....((....)). ( -47.80, z-score = -1.16, R) >droPer1.super_0 9811350 96 + 11822988 UGGUAUGUGGGUGUGGGUGAGUACCAGCAGCCGGUGGCCGCCAGCUGCUGGCCGUAAGCGGCGGC------AGCAGUGGCCCCUAAGCCGUAUGGGAACCCG .(((....((.(((.(((....))).))).))((.((((((..(((((((.(((....)))))))------))).))))))))...)))....((....)). ( -47.80, z-score = -1.16, R) >droWil1.scaffold_181108 1137323 102 - 4707319 GGAUAAGUGGGUGUCGGUGAAUACCAGCAGCCAGUGGCGGCCAAUUGCUGGCCAUAGGCGGCAGCAGCUGCAGCAGCAGCGCCCACGCCAUACGGAUAUCCG (((((.((((((((((((....)))....(((.((((((((.....))).))))).))))))....(((((....))))))))))).((....)).))))). ( -47.50, z-score = -2.40, R) >droVir3.scaffold_13047 3565788 101 + 19223366 GGAUAGGUAGGCGUCGGCGAAUACCAGCACCCAGUGGCGGCCAACUGCUGCCCAUAAGCGGCAGCGGCGGCAGCAGCAGCGCUCACGCCAUACGGAAAGCC- .....((((..((....))..)))).((..((.(((((((((..(((((((((....).)))))))).)))(((......)))..))))))..))...)).- ( -41.20, z-score = -0.48, R) >droGri2.scaffold_14830 4348559 102 - 6267026 GGAUAAGUGGGCGUCGGUGAAUACCAGCACCCAGUGGCUGCCAGCUGCUGACCAUAUGCGGCAGCUGCGGCUGCGGCAGCACUAACGCCGUACGGAAAACCG ......((((((((..(((....((.(....).(..(((((.((((((((........)))))))))))))..)))...)))..))))).)))((....)). ( -44.20, z-score = -1.41, R) >consensus GGGUAGGUGGGCGUCGGUGAGUACCAGCAGCCAGUGGCGGCCAGCUGCUGGCCGUAAGCGGCGGC_GC___AGCAGCAGCGCCCACGCCAUACGGGAAACCG ......((((((((.(((........((.....)).(((((..(((((((........))))))).((....)).)))))))).))))).)))((....)). (-25.82 = -26.21 + 0.40)

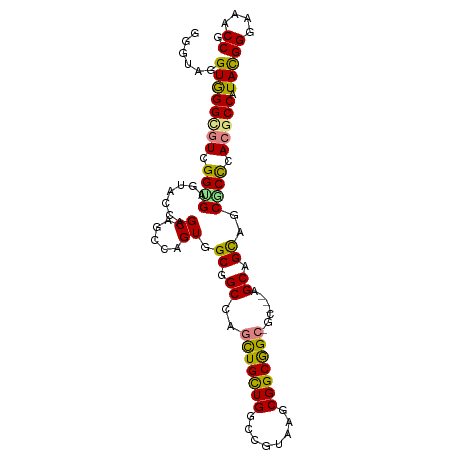
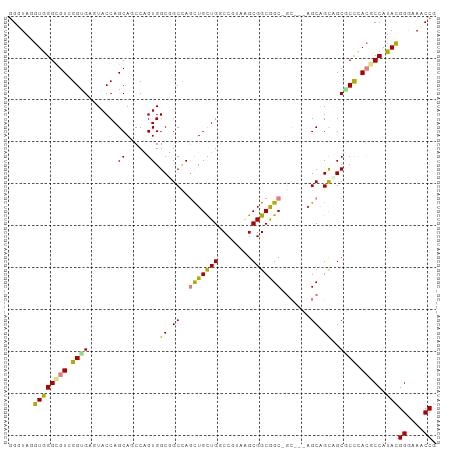
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:55 2011