
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,908,303 – 23,908,418 |
| Length | 115 |
| Max. P | 0.827586 |

| Location | 23,908,303 – 23,908,405 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 79.57 |
| Shannon entropy | 0.40379 |
| G+C content | 0.42712 |
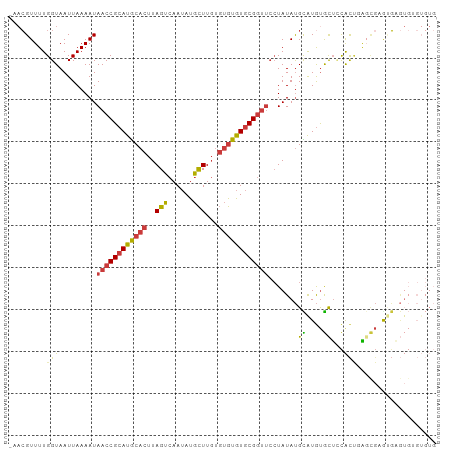
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -16.25 |
| Energy contribution | -16.59 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23908303 102 - 27905053 CAACGUUUUGGUAAUUAAAAUAACCGCAUGCACUUAGUCAAUAUGCUUGUGUGUGUGCGGUUCCUAUAUGCAUGUGCUCCACCGAGCGAGUGAGUGUGUGUG .....................((((((((((((..(((......)))...))))))))))))..(((((((((.(((((....))))).....))))))))) ( -31.10, z-score = -1.70, R) >droSim1.chr3R 23608310 101 - 27517382 -AACGUUUUGGUAAUUAAAAUAACCGCAUGCACUUAGUCAAUAUGCUUGUGUGUGUGCGGUUCCUAUAUGCAUGUGCUCCACCGAGCGCGUGAGUGUGUGUG -....................((((((((((((..(((......)))...))))))))))))..(((((.(((((((((....))))))))).))))).... ( -35.50, z-score = -2.95, R) >droSec1.super_22 526858 101 - 1066717 -AACGUUUUGGUAAUUAAAAUAACCGCAUGCACUUAGUCAAUAUGCUUGUGUGUGUGCGGUUCCUAUAUGCAUGUGCUCCACUGAGCGCGUGAGUGGGUGUG -....................((((((((((((..(((......)))...))))))))))))(((((...(((((((((....))))))))).))))).... ( -37.70, z-score = -3.35, R) >droYak2.chr3R 6243200 101 + 28832112 -AACGUUUUGGUAAUUAAAAUAACCGCAUGCACUUAGUCAAUAUGCUUGUGUGUGUGCGGUUCCUAUAUGCAUGUGCUACACUCGCUGAAUGCGUGUGUGUG -...((....(((........((((((((((((..(((......)))...))))))))))))......)))....))(((((.(((.....))).))))).. ( -31.04, z-score = -1.78, R) >droEre2.scaffold_4820 6332533 101 + 10470090 -AACGUUUUGGUAAUUAAAAUAACCGCAUGCACUUAGUCAAUAUGCUUGUGUGUGUGCGGUUCCUAUAUGCGUGUGCUCCACUCGGUGAAGGAGUGUGUGUG -....................((((((((((((..(((......)))...)))))))))))).......(((..((((((..........))))))..))). ( -31.00, z-score = -2.05, R) >droAna3.scaffold_13340 22049501 90 - 23697760 -AACGUUUUGGUAAUUAAAAUAACCGCAUGCACUUAGUCAAUAUGCUUGUGUGUGUGCGGUUCCUAUACGUGUACACUAUACACAC---ACGUA-------- -.((((...............((((((((((((..(((......)))...)))))))))))).......(((((.....)))))..---)))).-------- ( -26.50, z-score = -1.82, R) >droMoj3.scaffold_6540 14994605 87 + 34148556 -ACGGUUUUGGUAAUUAAAAUAACCGCAUACACUUAGUCAAAAUAUUUCCCCAUAUACG-----UAUAUGUGUGUGUGUCGUUGUGUGUGGCU--------- -.(((((..............)))))(((((((...(.((...........((((((((-----....)))))))))).)...)))))))...--------- ( -18.44, z-score = -0.30, R) >consensus _AACGUUUUGGUAAUUAAAAUAACCGCAUGCACUUAGUCAAUAUGCUUGUGUGUGUGCGGUUCCUAUAUGCAUGUGCUCCACUGAGCGAGUGAGUGUGUGUG ....((((((.....))))))((((((((((((..(((......)))...))))))))))))........(((.((((......)))).))).......... (-16.25 = -16.59 + 0.33)

| Location | 23,908,325 – 23,908,418 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 82.16 |
| Shannon entropy | 0.34784 |
| G+C content | 0.43252 |
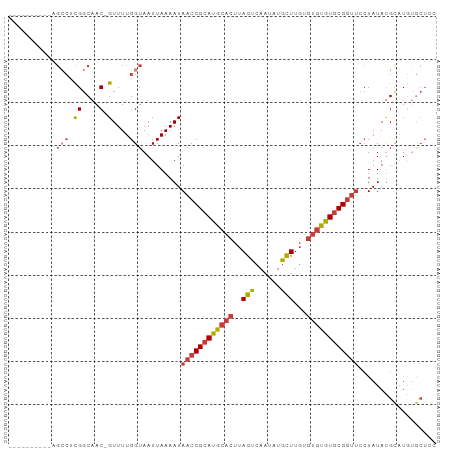
| Mean single sequence MFE | -24.34 |
| Consensus MFE | -16.76 |
| Energy contribution | -17.31 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.536823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23908325 93 - 27905053 -----AGCCUCGGCUCGGCAAC-GUUUUGGUAAUUAAAAUAACCGCAUGCACUUAGUCAAUAUGCUUGUGUGUGUGCGGUUCCUAUAUGCAUGUGCUCC -----......(((.(((((..-((((((.....))))))((((((((((((..(((......)))...))))))))))))......))).)).))).. ( -26.50, z-score = -1.22, R) >droSim1.chr3R 23608332 88 - 27517382 ----------AGCCUCGGCAAC-GUUUUGGUAAUUAAAAUAACCGCAUGCACUUAGUCAAUAUGCUUGUGUGUGUGCGGUUCCUAUAUGCAUGUGCUCC ----------.(((.((....)-)....))).........((((((((((((..(((......)))...)))))))))))).......((....))... ( -26.50, z-score = -2.05, R) >droSec1.super_22 526880 88 - 1066717 ----------AGCCUCGGCAAC-GUUUUGGUAAUUAAAAUAACCGCAUGCACUUAGUCAAUAUGCUUGUGUGUGUGCGGUUCCUAUAUGCAUGUGCUCC ----------.(((.((....)-)....))).........((((((((((((..(((......)))...)))))))))))).......((....))... ( -26.50, z-score = -2.05, R) >droYak2.chr3R 6243222 88 + 28832112 ----------AGCCUCGGCAAC-GUUUUGGUAAUUAAAAUAACCGCAUGCACUUAGUCAAUAUGCUUGUGUGUGUGCGGUUCCUAUAUGCAUGUGCUAC ----------.(((.((....)-)....))).........((((((((((((..(((......)))...)))))))))))).......((....))... ( -26.50, z-score = -1.74, R) >droEre2.scaffold_4820 6332555 88 + 10470090 ----------AGCCUCGGCAAC-GUUUUGGUAAUUAAAAUAACCGCAUGCACUUAGUCAAUAUGCUUGUGUGUGUGCGGUUCCUAUAUGCGUGUGCUCC ----------......((((((-((...((..........((((((((((((..(((......)))...)))))))))))))).....)))).)))).. ( -26.80, z-score = -2.04, R) >droAna3.scaffold_13340 22049512 98 - 23697760 UGCCCGACUGGGCCUCGGCAAC-GUUUUGGUAAUUAAAAUAACCGCAUGCACUUAGUCAAUAUGCUUGUGUGUGUGCGGUUCCUAUACGUGUACACUAU .((((....))))...(...((-((...((..........((((((((((((..(((......)))...))))))))))))))...))))...)..... ( -31.00, z-score = -1.77, R) >droMoj3.scaffold_6540 14994625 72 + 34148556 ---------------UGCCUACGGUUUUGGUAAUUAAAAUAACCGCAUACACUUAGUCAAAAUAUUUCCCCAUAUACG-----UAUAUGUGU------- ---------------((((.........))))...........((((((.((.((...................)).)-----).)))))).------- ( -6.61, z-score = 1.41, R) >consensus __________AGCCUCGGCAAC_GUUUUGGUAAUUAAAAUAACCGCAUGCACUUAGUCAAUAUGCUUGUGUGUGUGCGGUUCCUAUAUGCAUGUGCUCC .................((....((((((.....))))))((((((((((((..(((......)))...)))))))))))).......))......... (-16.76 = -17.31 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:54 2011