
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,901,910 – 23,902,006 |
| Length | 96 |
| Max. P | 0.806038 |

| Location | 23,901,910 – 23,902,006 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 73.76 |
| Shannon entropy | 0.50382 |
| G+C content | 0.48329 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -13.59 |
| Energy contribution | -12.99 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.806038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
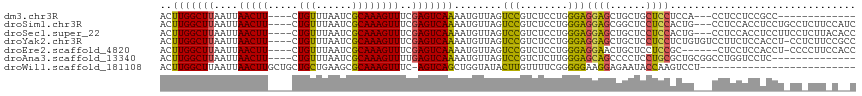
>dm3.chr3R 23901910 96 + 27905053 ACUUGGCUUAAUUAACUU----CUGUUUAAUCGCAAAGUUUCGAGUCAAAAUGUUAGUCCGUCUCCUGGGAGGAGCUGCUGCUCCUCCA---CCUCCUCCGCC------------- ..(((((((....(((((----.(((......))))))))..)))))))...................((((((((....)))))))).---...........------------- ( -26.70, z-score = -2.60, R) >droSim1.chr3R 23601824 109 + 27517382 ACUUGGCUUAAUUAACUU----CUGUUUAAUCGCAAAGUUUCGAGUCAAAAUGUUAGUCCGUCUCCUGGGAGGAGCGGCUCCUCCACUG---CCUCCACCUCCUGCCUCUUCCAUC ..(((((((....(((((----.(((......))))))))..))))))).(((..((...((.....((((((.(.(((.........)---)).)..))))))))..))..))). ( -24.10, z-score = -0.83, R) >droSec1.super_22 519515 109 + 1066717 ACUUGGCUUAAUUAACUU----CUGUUUAAUCGCAAAGUUUCGAGUCAAAAUGUUAGUCCGUCUCCUGGGAGGAGCUGCUCCUCCACUG---CCUCCACCUCCUUCCUCUUACACC ..(((((((....(((((----.(((......))))))))..)))))))...........((.....((((((((.((...(......)---....)).))))))))....))... ( -21.80, z-score = -0.86, R) >droYak2.chr3R 6236081 111 - 28832112 ACUUGGCUUAAUUAACUU----CUGUUUAAUCGCAAAGUUUCGAGUCAAAAUGUUAGUCCGUCUCCUGGGAGGAGCUGCUCCUCCUCUGUGUCCUUCUCCACCU-CCUCUUCCGCC ..(((((((....(((((----.(((......))))))))..)))))))...........(.(.(..((((((((...))))))))..).).)...........-........... ( -21.90, z-score = -0.82, R) >droEre2.scaffold_4820 6325823 105 - 10470090 ACUUGGCUUAAUUAACUU----CUGUUUAAUCGCAAAGUUUCGAGUCAAAAUGUUAGUCCGUCUCCUGGGAGGAACUGCUCCUCCGC------CUCCUCCACCU-CCCCUUCCACC ..(((((((....(((((----.(((......))))))))..)))))))...................((((((...((......))------.))))))....-........... ( -20.80, z-score = -1.22, R) >droAna3.scaffold_13340 22043088 98 + 23697760 ACUUGGCUUAAUUAACUU----CUGUUUAAUCGCAAAGUUUUGAGUCAAAAUGUUAGUCCGUCUCUUGGGAGCAGCCCCUCCUGCGCUGCGGCCUGGUCCUC-------------- ..(((((((((..(((((----.(((......)))))))))))))))))....((((.((((..(.((((((......)))))).)..)))).)))).....-------------- ( -25.30, z-score = -0.88, R) >droWil1.scaffold_181108 1128158 89 + 4707319 ACUUGGCUUAAUUAACUUGCUGCUGCUGAAGCGCAAAGUUUC-AGUCAGCUGGUAUACUUGUUUUCGGGGGAAGGAGAAUACCAAGUCCU-------------------------- ....(((((............(((((((((((.....)))))-)).)))).(((((.(((.((((....)))).))).))))))))))..-------------------------- ( -30.40, z-score = -2.34, R) >consensus ACUUGGCUUAAUUAACUU____CUGUUUAAUCGCAAAGUUUCGAGUCAAAAUGUUAGUCCGUCUCCUGGGAGGAGCUGCUCCUCCGCUG___CCUCCUCCUCCU_CC_CUU_C__C ..(((((((....(((((.....(((......))))))))..)))))))........(((........)))((((......))))............................... (-13.59 = -12.99 + -0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:51 2011