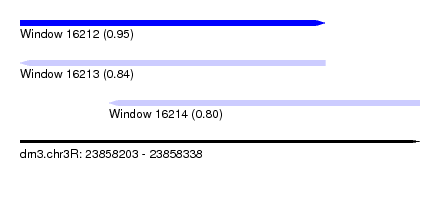
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,858,203 – 23,858,338 |
| Length | 135 |
| Max. P | 0.949326 |

| Location | 23,858,203 – 23,858,306 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.76 |
| Shannon entropy | 0.40026 |
| G+C content | 0.58686 |
| Mean single sequence MFE | -42.98 |
| Consensus MFE | -23.09 |
| Energy contribution | -24.60 |
| Covariance contribution | 1.51 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
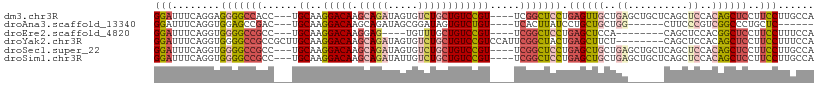
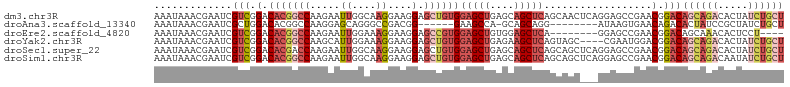
>dm3.chr3R 23858203 103 + 27905053 GGAUUUCAGGAGGGGCCACC---UGCAAGGACAAGCAGAUAGUGUCUGCUGUCCGU----UCGGCUCCUGAGUUGCUGAGCUGCUCAGCUCCACAGCUCCUUCCUUGCCA ((.....((((((((((...---.((..(((((.((((((...)))))))))))))----..)))))).((((((..(((((....)))))..))))))..))))..)). ( -45.50, z-score = -2.17, R) >droAna3.scaffold_13340 21997807 91 + 23697760 GGAUUUCAGGUGGAGCCGAC---UGCAAGGACAAGCAGAUAGCGGAUAGUGUCUGU----UCACUUAUCCUGCUGCUGG------CUUCCCGUCGGCCCUGCUC------ ........((..(.((((((---.....(((..(((((.((((((((((((.....----.))).))))).)))))).)------))))).)))))).)..)).------ ( -34.20, z-score = -1.22, R) >droEre2.scaffold_4820 6287331 91 - 10470090 GGAUUUCAGGUGGGGCCGCC---UGCAAGGACAAGGAG----UGUUUGCUGUCCGU----UCGGCUCCUGAGCUCCA--------CAGCUCCACGGCUCCUUCCUUUCCA (((...((((((....))))---)).(((((..(((((----(((..(((((..((----((((...))))))...)--------))))...)).)))))))))))))). ( -37.10, z-score = -1.52, R) >droYak2.chr3R 6192286 102 - 28832112 GGAUUUCAGGUGGGGCCGCCGCUUGCAAGGACAAGCAGAUAGUGUCUGCUGUCCGUCCAUUCGGCUACUGAGCUUCU--------CAGCUCCACAGCUCCUUCCUUUCCA (((....(((.(((((.((((..((...(((((.((((((...)))))))))))...))..))))....(((((...--------.)))))....)))))..))).))). ( -44.10, z-score = -3.77, R) >droSec1.super_22 480872 103 + 1066717 GGAUUUCAGGUGGGGCCGCC---UGCAAGGACAAGCAGAUAGUGUCUGCUGUCCGU----UCGGCUCCUGAGCUGCUGAGCUGCUCAGCUCCACAGCUCCUUCCUUGCCA ........((((((((((..---.((..(((((.((((((...)))))))))))))----.))))))).((((((..(((((....)))))..)))))).......))). ( -48.50, z-score = -2.64, R) >droSim1.chr3R 23559949 103 + 27517382 GGAUUUCAGGUGGGGCCGCC---UGCAAGGACAAGCAGAUAUUGUCUGCUGUCCGU----UCGGCUCCUGAGCUGCUGAGCUGCUCAGCUCCACAGCUCCUUCCUUGCCA ........((((((((((..---.((..(((((.((((((...)))))))))))))----.))))))).((((((..(((((....)))))..)))))).......))). ( -48.50, z-score = -3.03, R) >consensus GGAUUUCAGGUGGGGCCGCC___UGCAAGGACAAGCAGAUAGUGUCUGCUGUCCGU____UCGGCUCCUGAGCUGCUGA______CAGCUCCACAGCUCCUUCCUUGCCA (((........(((((((..........(((((.(((((.....)))))))))).......))))))).((((((..................))))))..)))...... (-23.09 = -24.60 + 1.51)
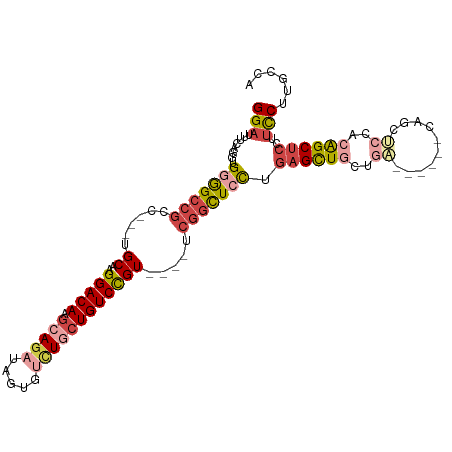
| Location | 23,858,203 – 23,858,306 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.76 |
| Shannon entropy | 0.40026 |
| G+C content | 0.58686 |
| Mean single sequence MFE | -40.22 |
| Consensus MFE | -22.12 |
| Energy contribution | -24.07 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
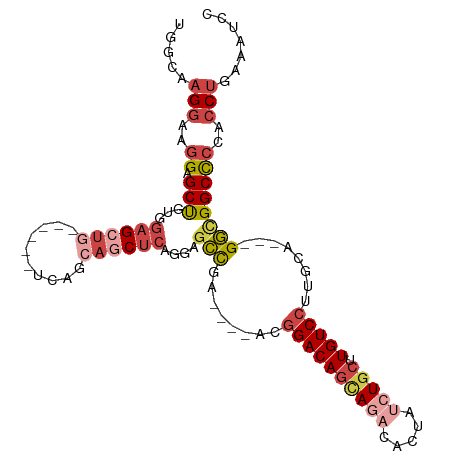
>dm3.chr3R 23858203 103 - 27905053 UGGCAAGGAAGGAGCUGUGGAGCUGAGCAGCUCAGCAACUCAGGAGCCGA----ACGGACAGCAGACACUAUCUGCUUGUCCUUGCA---GGUGGCCCCUCCUGAAAUCC .....((((..(((.(((.(((((....))))).))).))).((.((((.----..((((((((((.....))))).))))).....---..)))).))))))....... ( -39.50, z-score = -0.85, R) >droAna3.scaffold_13340 21997807 91 - 23697760 ------GAGCAGGGCCGACGGGAAG------CCAGCAGCAGGAUAAGUGA----ACAGACACUAUCCGCUAUCUGCUUGUCCUUGCA---GUCGGCUCCACCUGAAAUCC ------((.((((((((((((....------))....(((((((((((..----(.((.(.......))).)..)))))))).))).---))))))....))))...)). ( -33.60, z-score = -2.02, R) >droEre2.scaffold_4820 6287331 91 + 10470090 UGGAAAGGAAGGAGCCGUGGAGCUG--------UGGAGCUCAGGAGCCGA----ACGGACAGCAAACA----CUCCUUGUCCUUGCA---GGCGGCCCCACCUGAAAUCC .(((.(((..((.(((...(((((.--------...)))))....(((..----..((((((......----....)))))).....---))))))))..)))....))) ( -31.00, z-score = -0.15, R) >droYak2.chr3R 6192286 102 + 28832112 UGGAAAGGAAGGAGCUGUGGAGCUG--------AGAAGCUCAGUAGCCGAAUGGACGGACAGCAGACACUAUCUGCUUGUCCUUGCAAGCGGCGGCCCCACCUGAAAUCC .(((.(((..((.(((...(((((.--------...)))))....((((..((.(.((((((((((.....))))).))))).).))..)))))))))..)))....))) ( -43.00, z-score = -3.33, R) >droSec1.super_22 480872 103 - 1066717 UGGCAAGGAAGGAGCUGUGGAGCUGAGCAGCUCAGCAGCUCAGGAGCCGA----ACGGACAGCAGACACUAUCUGCUUGUCCUUGCA---GGCGGCCCCACCUGAAAUCC .....(((...(((((((.(((((....))))).))))))).((.((((.----..((((((((((.....))))).))))).....---..)))).)).)))....... ( -47.10, z-score = -2.63, R) >droSim1.chr3R 23559949 103 - 27517382 UGGCAAGGAAGGAGCUGUGGAGCUGAGCAGCUCAGCAGCUCAGGAGCCGA----ACGGACAGCAGACAAUAUCUGCUUGUCCUUGCA---GGCGGCCCCACCUGAAAUCC .....(((...(((((((.(((((....))))).))))))).((.((((.----..((((((((((.....))))).))))).....---..)))).)).)))....... ( -47.10, z-score = -2.98, R) >consensus UGGCAAGGAAGGAGCUGUGGAGCUG______UCAGCAGCUCAGGAGCCGA____ACGGACAGCAGACACUAUCUGCUUGUCCUUGCA___GGCGGCCCCACCUGAAAUCC .....(((..((.(((((.((((((..........))))))...............((((((((((.....))))).))))).........)))))))..)))....... (-22.12 = -24.07 + 1.95)

| Location | 23,858,233 – 23,858,338 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.45 |
| Shannon entropy | 0.43222 |
| G+C content | 0.53282 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -14.32 |
| Energy contribution | -16.63 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.795724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
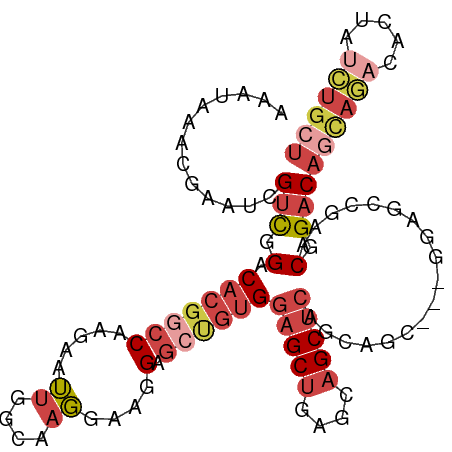
>dm3.chr3R 23858233 105 - 27905053 AAAUAAACGAAUCGUCGGACACGGCCAAGAAUUGGCAAGGAAGGAGCUGUGGAGCUGAGCAGCUCAGCAACUCAGGAGCCGAACGGACAGCAGACACUAUCUGCU ...........(((((((...(.((((.....)))).......(((.(((.(((((....))))).))).))).)...))).))))..((((((.....)))))) ( -34.30, z-score = -2.03, R) >droAna3.scaffold_13340 21997837 90 - 23697760 AAAUAAACGAAUCGCUGGACACGGCCAAGGAGCAGGGCCGACGG------GAAGCCA-GCAGCAGG--------AUAAGUGAACAGACACUAUCCGCUAUCUGCU .............(((((.(.(((((.........)))))...)------....)))-))(((.((--------(((.(((......)))))))))))....... ( -28.60, z-score = -2.05, R) >droEre2.scaffold_4820 6287361 93 + 10470090 AAAUAAACGAAUCGUCGGACACGGCCAAGAAUUGGAAAGGAAGGAGCCGUGGAGCUGUGGAGCUCA--------GGAGCCGAACGGACAGCAAACACUCCU---- ...........(((((((.(((((((.....((....))....).))))))(((((....))))).--------....))).))))...............---- ( -25.70, z-score = -0.69, R) >droYak2.chr3R 6192319 101 + 28832112 AAAUAAACGAAUCGUCGGACACGGCCAAGCAUUGGAAAGGAAGGAGCUGUGGAGCUGAGAAGCUCAGUAGC----CGAAUGGACGGACAGCAGACACUAUCUGCU .......((.....((((.(((((((.....((....))....).))))))(((((....))))).....)----))).....))...((((((.....)))))) ( -29.80, z-score = -1.61, R) >droSec1.super_22 480902 105 - 1066717 AAAUAAACGAAUCGUCGGACACGACCAAGAAUUGGCAAGGAAGGAGCUGUGGAGCUGAGCAGCUCAGCAGCUCAGGAGCCGAACGGACAGCAGACACUAUCUGCU .......((....((((....))))......(((((.......(((((((.(((((....))))).)))))))....))))).))...((((((.....)))))) ( -38.60, z-score = -3.31, R) >droSim1.chr3R 23559979 105 - 27517382 AAAUAAACGAAUCGUCGGACACGGCCAAGAAUUGGCAAGGAAGGAGCUGUGGAGCUGAGCAGCUCAGCAGCUCAGGAGCCGAACGGACAGCAGACAAUAUCUGCU ...........(((((((...(.((((.....)))).......(((((((.(((((....))))).))))))).)...))).))))..((((((.....)))))) ( -40.90, z-score = -3.91, R) >consensus AAAUAAACGAAUCGUCGGACACGGCCAAGAAUUGGCAAGGAAGGAGCUGUGGAGCUGAGCAGCUCAGCAGC___GGAGCCGAACGGACAGCAGACACUAUCUGCU .............(((.(.(((((((.....((....))....).))))))(((((....)))))..................).)))((((((.....)))))) (-14.32 = -16.63 + 2.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:50 2011