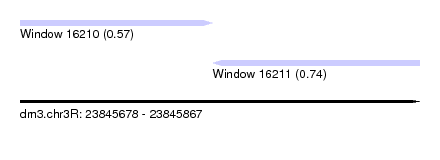
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,845,678 – 23,845,867 |
| Length | 189 |
| Max. P | 0.735082 |

| Location | 23,845,678 – 23,845,769 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 72.97 |
| Shannon entropy | 0.51960 |
| G+C content | 0.60686 |
| Mean single sequence MFE | -38.87 |
| Consensus MFE | -20.49 |
| Energy contribution | -20.80 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
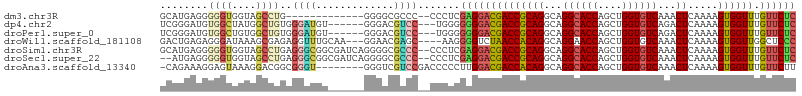
>dm3.chr3R 23845678 91 + 27905053 GCAUGAGGGGGUGGUAGCCUG-------------GGGGCGCCC--CCCUCGAGGACGACCGCAGGCAGGCACCAGCUGGUGUCAAACUCAAAAGUGGUUUGUUCUC ....(((((((.(...(((..-------------..))).)))--)))))((((((((((((((...((((((....))))))...)).....)))).)))))))) ( -42.80, z-score = -1.98, R) >dp4.chr2 22554513 97 - 30794189 UCGGGAUGUGGCUAUGGCUGUGGGAUGU------GGGACGUCC---UGGGGGGGACGACCGCAGGCAGGCACCAGCUGGUGUCAGACUCAAAAGUGGUUUGUUCUC ..((((...((((((.....((((.(((------((..(((((---(....)))))).)))))....((((((....))))))...))))...))))))...)))) ( -37.70, z-score = -1.27, R) >droPer1.super_0 9747597 97 - 11822988 UCGGGAUGUGGCUGUGGCUGUGGGAUGU------GGGACGUCC---UGGGGGGGACGACCGCAGGCAGGCACCAGCUGGUGUCAGACUCAAAAGUGGUUUGUUCUC ..((((...(((((((.((((....(((------((..(((((---(....)))))).))))).)))).)).))))).....(((((.((....))))))).)))) ( -36.40, z-score = -0.57, R) >droWil1.scaffold_181108 1066238 99 + 4707319 GACUGAGAGGGAUAAAGCGAGAGGUUUGCAA---GGAACGAGC----AAGGGGUCUAACCACAGGCAGGAACCAGCUGGUGUCAAACUCAAAAGUGGUUGGCUCCC ..((....))(.((((.(....).)))))..---.........----..((((.((((((((.(((.(....).)))(((.....))).....)))))))))))). ( -25.10, z-score = 0.73, R) >droSim1.chr3R 23547485 104 + 27517382 GCAUGAGGGGGUGGUAGCCUGAGGGCGGCGAUCAGGGGCGCCC--CCCUCGAGGACGACCGCAGGCAGGCACCAGCUGGUGUCAAACUCAAAAGUGGUUUGUUCUC ....(((((((..((..(((((.........))))).))..))--)))))((((((((((((((...((((((....))))))...)).....)))).)))))))) ( -47.00, z-score = -1.55, R) >droSec1.super_22 468330 102 + 1066717 --AUGAGGGGGUGGUAGCCUGAGGGCGGCGAUCAGGGGCGCCC--CCCUCGAGGACGACCGCAGGCAGGCACCAGCUGGUGUCAAACUCAAAAGUGGUUUGUUCUC --..(((((((..((..(((((.........))))).))..))--)))))((((((((((((((...((((((....))))))...)).....)))).)))))))) ( -47.00, z-score = -1.92, R) >droAna3.scaffold_13340 21984827 97 + 23697760 -CAGAAAGGAGUAAAGGACGGCGGGU--------GGGUCGUCCGACCCCCUUGGACGACCACAGGCAGGCACCAGCUGGUGUCAAACUCAAAAGUGGUUUGUUCUU -............((((((.((..((--------((.((((((((.....))))))))))))..)).((((((....))))))((((.((....)))))))))))) ( -36.10, z-score = -1.25, R) >consensus GCAUGAGGGGGUGGUAGCCGGAGGGUGG______GGGACGCCC__CCCUCGAGGACGACCGCAGGCAGGCACCAGCUGGUGUCAAACUCAAAAGUGGUUUGUUCUC ........((((....))))..............................((((((((((((((...((((((....))))))...)).....)))))).)))))) (-20.49 = -20.80 + 0.31)
| Location | 23,845,769 – 23,845,867 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.23 |
| Shannon entropy | 0.45512 |
| G+C content | 0.30402 |
| Mean single sequence MFE | -18.91 |
| Consensus MFE | -9.38 |
| Energy contribution | -10.77 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
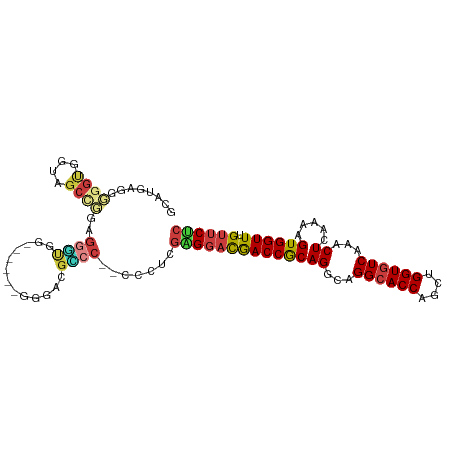
>dm3.chr3R 23845769 98 - 27905053 AAAUGUAAAGUUCAUGUUUAUAUUUUGGCUAAUG--UUUUUUCUAGUCGAGUAGUCGAGUGUGCUGGACAAAAUAAUAGAAA-AAGUGGCCGAGAAAAAAG--- ...((((((.......))))))(((((((((...--(((((((((........(((.((....)).))).......))))))-))))))))))))......--- ( -22.26, z-score = -2.79, R) >droWil1.scaffold_181108 1066337 99 - 4707319 AAAUUUAAACUUUAUGUUUAUAUUUUGGCUAAU---GUUUUCUAUUUUCUAGAGCUGGGUUAUCCAACGACAACAACAAAAAAUGAAAGAUGGGUUAAAAAU-- ...(((.(((((....(((((.(((((.....(---(((.((((.....))))(.(((.....))).)))))....))))).)))))....))))).)))..-- ( -13.10, z-score = 0.16, R) >droSim1.chr3R 23547589 98 - 27517382 AAAUGUAAAGUUCAUGUUUAUAUUUUGGCUAAUG--UUUUUUCUAGUCGAGUAGUCGAGUGUGCUGGACAAAAUAAUAGAAA-AAGUGGCCGAGAAAAAAG--- ...((((((.......))))))(((((((((...--(((((((((........(((.((....)).))).......))))))-))))))))))))......--- ( -22.26, z-score = -2.79, R) >droSec1.super_22 468432 98 - 1066717 AAAUGUAAAGUUCAUGUUUAUAUUUUGGCUAAUG--UUUUUUCUAGUCGAGUAGUCGAGUGUGCUGGACAAAAUAAUAGAAA-AAGUGGCCGAGAAAAAAG--- ...((((((.......))))))(((((((((...--(((((((((........(((.((....)).))).......))))))-))))))))))))......--- ( -22.26, z-score = -2.79, R) >droYak2.chr3R 6179058 103 + 28832112 AAAUGUAAAGUUCAUGUUUAUAUUUUGGCUAAUGUUUUUUUUCUAGUCGAGCAGUCGAGUGUGCUGGACAAAAUAAUAGAAA-AAGUAGCUGAGAAAAAAAAAG ...((((((.......))))))(((..((((.....((((((((((((.((((........)))).))).......))))))-)))))))..)))......... ( -20.71, z-score = -1.59, R) >droEre2.scaffold_4820 6274706 98 + 10470090 AAAUGUAAAGUUCAUGUUUAUAUUUUGGCUAAUG--UUUUUUCUAGUCGAGUAGUCGAGUGUGCUGGACAAAAUAAUAGAAA-AAGUGGCCGAGAAAAAAG--- ...((((((.......))))))(((((((((...--(((((((((........(((.((....)).))).......))))))-))))))))))))......--- ( -22.26, z-score = -2.79, R) >droMoj3.scaffold_6540 14918760 89 + 34148556 AAAUCGAAAGUUCAUGUUUAUAUUUUGGCUAAGG--GCUUUGCUAGUUGCAUCCACAA---------AAAAAAUAAUACAAAUAAAAUAACGAAAAUAUA---- ...((((((((((..(((........)))...))--)))))((.....))........---------.......................))).......---- ( -9.50, z-score = 0.33, R) >consensus AAAUGUAAAGUUCAUGUUUAUAUUUUGGCUAAUG__UUUUUUCUAGUCGAGUAGUCGAGUGUGCUGGACAAAAUAAUAGAAA_AAGUGGCCGAGAAAAAAG___ ......................(((((((((........((((((........(((.((....)).))).......))))))....)))))))))......... ( -9.38 = -10.77 + 1.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:48 2011