
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,839,430 – 23,839,530 |
| Length | 100 |
| Max. P | 0.862907 |

| Location | 23,839,430 – 23,839,530 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.54 |
| Shannon entropy | 0.43905 |
| G+C content | 0.45870 |
| Mean single sequence MFE | -26.76 |
| Consensus MFE | -19.45 |
| Energy contribution | -18.84 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.862907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23839430 100 + 27905053 -------AAACAACAAACGGUGGCAGGAGUGAGUGGCUCCGGAACUUUAGUGAUUAUUACUGACGUGGUUACGUGACUGUUUUGGAAAGUGUCG-AAAAUGCGGCACG -------......((((((((.((.(((((.....))))).........((((((((.......)))))))))).)))).))))....((((((-......)))))). ( -30.40, z-score = -1.92, R) >droSim1.chr3R 23541326 100 + 27517382 -------AAACAACAAACGGUGGCAGGACUGAGUGGCUCCGGAACUUUAGUGAUUAUUACUGACGUGGUUACGUGACUGUUUUGGAAAGUGUCG-AAAAUGCGGCACG -------......((((((((.((.(((((....)).))).........((((((((.......)))))))))).)))).))))....((((((-......)))))). ( -26.70, z-score = -0.75, R) >droSec1.super_22 462215 100 + 1066717 -------AAACAACAAACGGUGGCAGGACUGAGUGGCUCCGGAACUUUAGUGAUUAUUACUGACGUGGUUACGUGACUGUUUUGGAAAGUGUCG-AAAAUGCGGCACG -------......((((((((.((.(((((....)).))).........((((((((.......)))))))))).)))).))))....((((((-......)))))). ( -26.70, z-score = -0.75, R) >droYak2.chr3R 6172480 100 - 28832112 -------AAAAGAAGAACGGUGGCAGGAGUGAGUGGCUCCGGAACUUUAGUGAUUAUUACUGACGUGGUUACGUGACUGUUUUGGAAAGUGUCG-AAAAUGCGGCACG -------......((((((((.((.(((((.....))))).........((((((((.......)))))))))).)))))))).....((((((-......)))))). ( -31.80, z-score = -2.94, R) >droEre2.scaffold_4820 6268348 99 - 10470090 -------AAACAACAA-CGGUGGCAGGACUGAGUGGCUCCGGAACUUUAGUGAUUAUUACUGACGUGGUUACGUGACUGUUUUGGAAAGUGUCG-AAAAUGCGGCACG -------...(((.((-((((.((.(((((....)).))).........((((((((.......)))))))))).)))))))))....((((((-......)))))). ( -27.60, z-score = -1.06, R) >dp4.chr2 22548260 91 - 30794189 ------------UUACAUGG-GGCAGA---GAGUGGCUCCGAAACUUUAGUGAUUAUUACUGACGUGGUUACGUGACUGUUUUGGAAAGUGUCG-AAAAUGCGGCACG ------------.....(..-(((((.---..(((((..((.....(((((((...)))))))))..)))))....)))))..)....((((((-......)))))). ( -24.70, z-score = -1.24, R) >droPer1.super_0 9741384 91 - 11822988 ------------UUACAUGG-GGCAGA---GAGUGGCUCCGAAACUUUAGUGAUUAUUACUGACGUGGUUACGUGACUGUUUUGGAAAGUGUCG-AAAAUGCGGCACG ------------.....(..-(((((.---..(((((..((.....(((((((...)))))))))..)))))....)))))..)....((((((-......)))))). ( -24.70, z-score = -1.24, R) >droWil1.scaffold_181108 1054863 103 + 4707319 ----GAGCAAGCAGCAGUAAGGAGUUGGUAUGGUGUAACUGAAACUUUAGUGAUUAUUACUGACGUGGUUACGUGACUGUUUUGGAAAGUGUCG-AAAAUGCGGCACA ----....((((((((((((...((..(((((((...(((((....))))).)))).)))..))....)))).)).))))))......((((((-......)))))). ( -26.40, z-score = -0.82, R) >droMoj3.scaffold_6540 14909918 91 - 34148556 -----------------GGCUGGGUAGAUUGUAGGUAGCUGAAACUUUAGUGAUUAUUACUAACGUGGUUACGUGACUGUUUUGGAAAGUGCCGGAAAAUGCGGCACA -----------------..(..((.......((.((((((...((.(((((((...))))))).)))))))).)).....))..)...((((((.......)))))). ( -23.00, z-score = -0.84, R) >droGri2.scaffold_14830 4274375 88 + 6267026 ------------------GGGGGGU-GUUUGUAAGUAGCAGAAACUUUAGUGAUUAUUACUAACGUGGUUACGUGACUGUUUUGGAAAGUGCCG-AAAAUGCGGCACG ------------------..(((((-.(((((.....))))).)))))(((.((.(((((....)))))...)).)))..........((((((-......)))))). ( -22.80, z-score = -1.22, R) >droVir3.scaffold_13047 3487423 107 - 19223366 AAGCAAUGGCAUCGGCCAGGAGGGUAGACUGUUGGUAGCUGAAACUUUAGUGAUUAUUACUAACGUGGUUACGUGACUGUUUUGGAAAGUGCCG-AAAAUGUGGCACG ..((....))....(((((.((......)).)))))..(..((((...(((.((.(((((....)))))...)).)))))))..)...((((((-......)))))). ( -29.60, z-score = -1.02, R) >consensus _______AAA__ACAAACGGUGGCAGGACUGAGUGGCUCCGAAACUUUAGUGAUUAUUACUGACGUGGUUACGUGACUGUUUUGGAAAGUGUCG_AAAAUGCGGCACG ......................................(((((((.(((((((((((.......)))))))).)))..)))))))...((((((.......)))))). (-19.45 = -18.84 + -0.61)
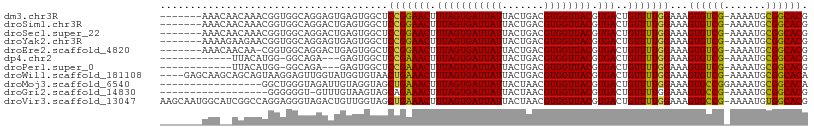
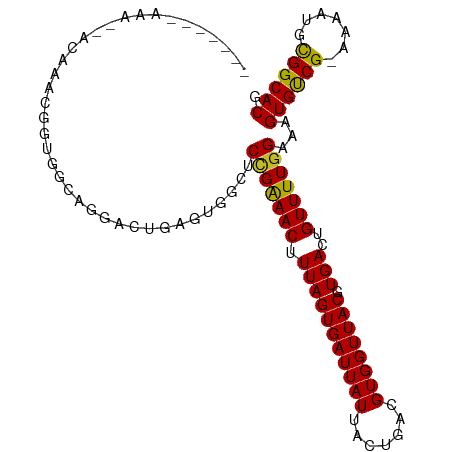

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:46 2011