| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,831,591 – 23,831,691 |
| Length | 100 |
| Max. P | 0.910956 |

| Location | 23,831,591 – 23,831,691 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 74.06 |
| Shannon entropy | 0.48978 |
| G+C content | 0.37666 |
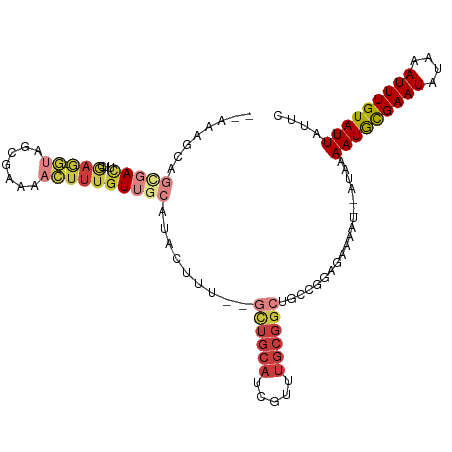
| Mean single sequence MFE | -26.26 |
| Consensus MFE | -10.88 |
| Energy contribution | -12.27 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
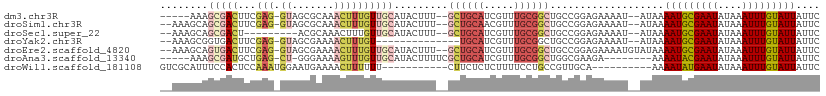
>dm3.chr3R 23831591 100 - 27905053 -----AAAGCGACUUCGAG-GUAGCGCAAACUUUGUUGCAUACUUU--GCUGCAUCGUUUGCGGCUGCCGGAGAAAAU--AUAAAAUGCGAAUAUAAAUUUGUAUUAUUC -----.......((((..(-(((((((((((..(((.(((.....)--)).)))..)))))).)))))))))).....--....(((((((((....))))))))).... ( -33.20, z-score = -3.67, R) >droSim1.chr3R 23533475 103 - 27517382 --AAAGCAGCGACUUCGAG-GUAGCGCAAACUUUGUUGCAUACUUU--GCUGCAACGUUUGCGGCUGCCGGAGAAAAU--AUAAAAUGCGAAUAUAAAUUUGUAUUAUUC --..........((((..(-(((((((((((.((((.(((.....)--)).)))).)))))).)))))))))).....--....(((((((((....))))))))).... ( -34.00, z-score = -3.37, R) >droSec1.super_22 454435 95 - 1066717 --AAAGCAGCGACU---------ACGCAAACUUUGUUGCAUACUUU--GCUGCAUCGUUUGCGGCUGCCGGAGAAAAU--AUAAAAUGCGAAUAUAAAUUUGUAUUAUUC --...(((((....---------.(((((((..(((.(((.....)--)).)))..))))))))))))..........--....(((((((((....))))))))).... ( -29.00, z-score = -3.20, R) >droYak2.chr3R 6164402 91 + 28832112 --AAAGCGGUGACUUCGAG-GUAGCGAAAACUUUGU--------------UGCAUCGUUUGCGGCUGCCGGAGAAAAU--AUAAAAUGCGAAUAUAAAUUUGUAUUAUUC --..(((.(..(...(((.-(((((((.....))))--------------))).))).)..).)))............--....(((((((((....))))))))).... ( -21.90, z-score = -1.17, R) >droEre2.scaffold_4820 6260452 105 + 10470090 --AAAGCAGUGACUUCGAG-GUAGCGAAAACUUUGUUGCAUACUUU--GCUGCAUCGUUUGCGGCUGCCGGAGAAAAUGUAUAAAAUGCGAAUAUAAAUUUGUAUUAUUC --...(((....((((..(-(((((..((((..(((.(((.....)--)).)))..))))...))))))))))....)))....(((((((((....))))))))).... ( -28.80, z-score = -1.96, R) >droAna3.scaffold_13340 21969914 95 - 23697760 -----AAAGCGAUGCUGAG-CU-GGGAAAAGUUUGUUGCAUACUUUUCGCUGCAUCGUUUGCGGCUGGCGAAGA--------AAAAUACGAAUAUAAAUUUGUAUUAUUC -----.(((((((((..((-(.-..(((((((.((...)).)))))))))))))))))))((.....))...((--------(.(((((((((....))))))))).))) ( -27.20, z-score = -2.21, R) >droWil1.scaffold_181108 1046157 89 - 4707319 GUCGCAUUUCCACUCCAAAUGGAAUGAAAACUUUUUU-----------CUUCUCUCUUUUCCUGCCGUUGCA----------AAAAUAUGAAUAUAAAUUUGUAUUAUUC ...(((.(((((.......))))).((((.....)))-----------)...................))).----------..((((..(((....)))..)))).... ( -9.70, z-score = -0.83, R) >consensus __AAAGCAGCGACUUCGAG_GUAGCGAAAACUUUGUUGCAUACUUU__GCUGCAUCGUUUGCGGCUGCCGGAGAAAAU__AUAAAAUGCGAAUAUAAAUUUGUAUUAUUC ........(((((.....................))))).........((((((.....))))))...................(((((((((....))))))))).... (-10.88 = -12.27 + 1.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:44 2011