| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,830,084 – 23,830,228 |
| Length | 144 |
| Max. P | 0.980099 |

| Location | 23,830,084 – 23,830,193 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.61 |
| Shannon entropy | 0.52453 |
| G+C content | 0.51912 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -15.33 |
| Energy contribution | -15.26 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.960903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
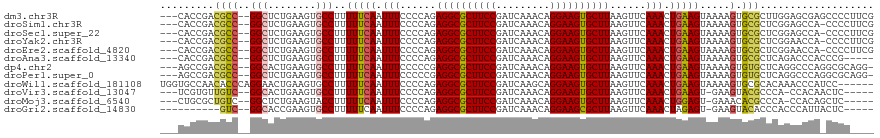
>dm3.chr3R 23830084 109 - 27905053 CCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGCGCUUGGAGCGAGCCCCUUCGCCCAACC------GCCACUUCGUAUAUCAGCGAGCA ......((((((((((.........))))))))))..((((...((((.(((.(((((((.((((.(((((....))))))))).)------).)))))..))).)))).)))). ( -41.70, z-score = -4.62, R) >droSim1.chr3R 23532014 108 - 27517382 CCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGCGCUCGGAGCCA-CCCCUUCGCCCAACC------GCCACUUCGUAUAUCAGCGAGCA ......((((((((((.........))))))))))..((((...((((.(((.(((((((...((.((..-.......))))...)------).)))))..))).)))).)))). ( -32.30, z-score = -2.50, R) >droSec1.super_22 452975 108 - 1066717 CCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGCGCUCGGAGCCA-CCCCUUCGCCCAACC------GCCACUUCGUAUAUCAGCGAGCA ......((((((((((.........))))))))))..((((...((((.(((.(((((((...((.((..-.......))))...)------).)))))..))).)))).)))). ( -32.30, z-score = -2.50, R) >droYak2.chr3R 6162919 114 + 28832112 CCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGCGCUCGGAACCA-CCCCUUCGCCCAACUUCGUCGCCCACUUCGUAUAUCAGCGAGCA ......((((((((((.........))))))))))..((((...((((.(((.(((((.((.((((....-..............))))..)).)))))..))).)))).)))). ( -31.37, z-score = -2.00, R) >droEre2.scaffold_4820 6255265 108 + 10470090 CCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGCGCUCGGAACCA-CCCCUUCGCCCAACC------GCCACUUCGUAUAUCAGCGAGCA ......((((((((((.........))))))))))..((((...((((.(((.(((((((...((.....-.........))...)------).)))))..))).)))).)))). ( -29.54, z-score = -2.03, R) >droAna3.scaffold_13340 21968300 107 - 23697760 CCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGCGCUCAGACCCA-CCCGCAGG-----ACUCUU--ACCACUUCGUAUAUCAGCCAGGA ..((..((((((((((.........)))))))))).........((((.(((.(((((.(...(((.((.-......))-----..))).--.))))))..))).))))...)). ( -26.70, z-score = -0.92, R) >dp4.chr2 22538621 112 + 30794189 CCCCCGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGUGCUCAGGCCCA-GGCGCAGGCCAACCCUCGU--ACCACUGCGUAUAUCAUCCAGCA ......((((((((((.........))))))))))..(((.....(((.(((..(((((((..(((....-(((....)))...))).))--).))))...))).)))...))). ( -30.40, z-score = -0.74, R) >droPer1.super_0 9731793 112 + 11822988 CCCCCGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGUGCUCAGGCCCA-GGCGCAGGCCAACCCUCGU--ACCACUGCGUAUAUCAUCCAGCA ......((((((((((.........))))))))))..(((.....(((.(((..(((((((..(((....-(((....)))...))).))--).))))...))).)))...))). ( -30.40, z-score = -0.74, R) >droWil1.scaffold_181108 1044315 101 - 4707319 CCCCAGAGGCGCUUCCGAUCAAGCAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGCGCACAAACCCA-UCCACAAU-------------UACACUAUCUUUAUCAUCUUGGA ..((((((((((((((.........))))))))))..........(((((....((((............-........-------------..))))..))))).....)))). ( -20.95, z-score = -0.97, R) >droVir3.scaffold_13047 3476769 100 + 19223366 CCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGU-GAAGUACGCCCA-CCACAACUCG-AAAAC-----------AUUGCUGGUUGAAGCAACAACG- .....(((((((((((.........))))))))...(.((((........)-))).).......-......))).-.....-----------.(((((......))))).....- ( -22.40, z-score = -0.57, R) >droMoj3.scaffold_6540 14896346 102 + 34148556 CCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGGAGU-GAAACACGCCCA-CCACAGCUCUGAAAAC-----------AUCAUUGACUGAGCCAAAGAGCG ......((((((((((.........))))))))))..((((....(((.((-(.....))))))-.....((((.......-----------..........))))....)))). ( -28.03, z-score = -2.02, R) >droGri2.scaffold_14830 4263877 99 - 6267026 CCCCAGAGGCGCUUCCGAUCAAACAGGAAGCGCUUAAGUUCAAACUAGAGU-GAAGUACACCCACCCAUUACUCC-AAAAU-----------AUUAUCGGUUGAAGCACCAG--- ......((((((((((.........))))))))))............((((-((.((......))...)))))).-.....-----------......(((......)))..--- ( -22.90, z-score = -2.68, R) >consensus CCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGCGCUCAGACCCA_CCCCUACGCC_A_CC______ACCACUUCGUAUAUCAGCGAGCA ......((((((((((.........))))))))))................................................................................ (-15.33 = -15.26 + -0.08)
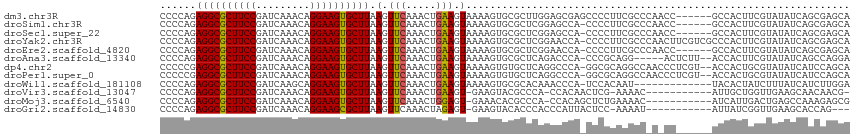
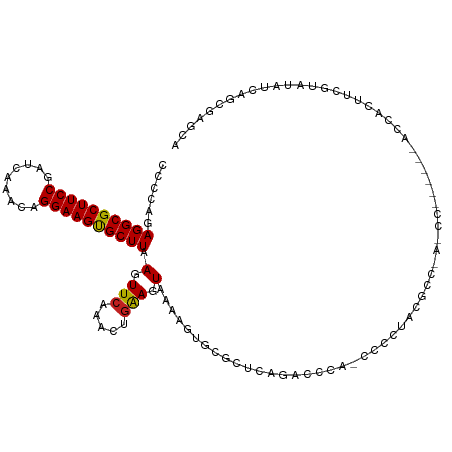
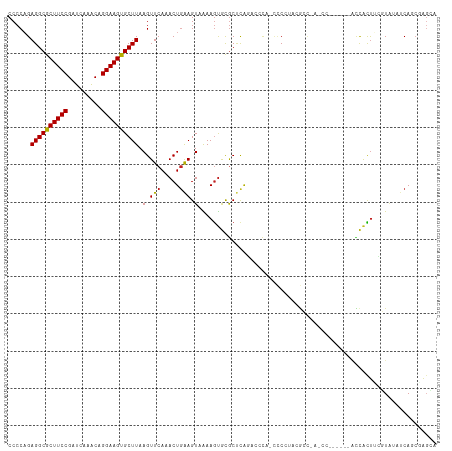
| Location | 23,830,114 – 23,830,228 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.22 |
| Shannon entropy | 0.34210 |
| G+C content | 0.51807 |
| Mean single sequence MFE | -35.91 |
| Consensus MFE | -22.41 |
| Energy contribution | -21.99 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
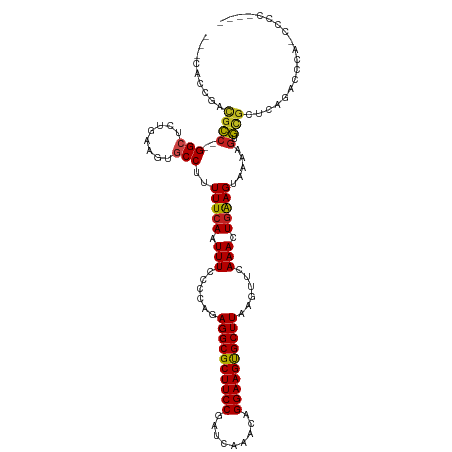
>dm3.chr3R 23830114 114 - 27905053 ---CACCGACGCC--GGCUCUGAAGUGCCUUUUUCAAUUUCCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGCGCUUGGAGCGAGCCCCUUCG ---.......((.--.(((((.((((((((((((((.(((......((((((((((.........))))))))))......))).)))...))))).)))))))))))..))....... ( -37.30, z-score = -2.05, R) >droSim1.chr3R 23532044 113 - 27517382 ---CACCGACGCC--GGCUCUGAAGUGCCUUUUUCAAUUUCCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGCGCUCGGAGCCA-CCCCUUCG ---..........--((((((((.((((((((((((.(((......((((((((((.........))))))))))......))).)))...))))).)))))))))))).-........ ( -40.90, z-score = -3.84, R) >droSec1.super_22 453005 113 - 1066717 ---CACCGACGCC--GGCUCUGAAGUGCCUUUUUCAAUUUCCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGCGCUCGGAGCCA-CCCCUUCG ---..........--((((((((.((((((((((((.(((......((((((((((.........))))))))))......))).)))...))))).)))))))))))).-........ ( -40.90, z-score = -3.84, R) >droYak2.chr3R 6162955 113 + 28832112 ---CACCGACGCC--GGCUCUGAAGUGCCUUUUUCAAUUUCCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGCGCUCGGAACCA-CCCCUUCG ---..........--((.(((((.((((((((((((.(((......((((((((((.........))))))))))......))).)))...))))).))))))))).)).-........ ( -34.30, z-score = -2.27, R) >droEre2.scaffold_4820 6255295 113 + 10470090 ---CACCGACGCC--GGCUCUGAAGUGCCUUUUUCAAUUUCCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGCGCUCGGAACCA-CCCCUUCG ---..........--((.(((((.((((((((((((.(((......((((((((((.........))))))))))......))).)))...))))).))))))))).)).-........ ( -34.30, z-score = -2.27, R) >droAna3.scaffold_13340 21968333 109 - 23697760 ---CACCGACGCC--GGCUCUGAAGUGCCUUUUUCAAUUUCCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGCGCUCAGACCCACCCG----- ---..........--((.(((((.((((((((((((.(((......((((((((((.........))))))))))......))).)))...))))).))))))))).)).....----- ( -35.00, z-score = -3.29, R) >dp4.chr2 22538655 113 + 30794189 ---AGCCGACGCC--GGCACUGAAGUGCCUUUUUCAAUUUCCCCCGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGUGCUCAGGCCCAGGCGCAGG- ---..((..((((--(((.((((.(..(((((((((.(((......((((((((((.........))))))))))......))).)))...))))).)..))))))))..))))..))- ( -42.40, z-score = -2.79, R) >droPer1.super_0 9731827 113 + 11822988 ---AGCCGACGCC--GGCUCUGAAGUGCCUUUUUCAAUUUCCCCCGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGUGCUCAGGCCCAGGCGCAGG- ---..((..((((--(((.((((.(..(((((((((.(((......((((((((((.........))))))))))......))).)))...))))).)..))))))))..))))..))- ( -42.40, z-score = -2.67, R) >droWil1.scaffold_181108 1044343 113 - 4707319 UGGUGCCAACACCCAGGAACUGAAGUGCCUUUUUCAAUUUCCCCAGAGGCGCUUCCGAUCAAGCAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGCGCACAAACCCAUCC------ ..((((..........((.(.(((((((((((............))))))))))).).))..(((.....(((((.(((....))).)))))....)))))))..........------ ( -29.90, z-score = -1.39, R) >droVir3.scaffold_13047 3476797 107 + 19223366 ---UCGUGUUGUC--GGCACUGAAGUGCCUUUUUCAAUUUCCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGU-GAAGUACGCCCA-CCACAACUC----- ---....(((((.--((..(.(((((((((((............))))))))))).)........((..((((((.(.(((.....))).)-.))))))..)).-)))))))..----- ( -32.50, z-score = -2.55, R) >droMoj3.scaffold_6540 14896376 107 + 34148556 ---CUGCGCUGUC--GGCUCUGAAGUACCUUUUUCAAUUUCCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGGAGU-GAAACACGCCCA-CCACAGCUC----- ---....(((((.--(((..(((((......))))).(((((((((((((((((((.........)))))))))).........)))).).-))))...)))..-..)))))..----- ( -33.20, z-score = -2.12, R) >droGri2.scaffold_14830 4263903 101 - 6267026 ----------GUC--GGCACCGAAGUGCCUUUUUCAAUUUCCCCAGAGGCGCUUCCGAUCAAACAGGAAGCGCUUAAGUUCAAACUAGAGU-GAAGUACACCCACCCAUUACUC----- ----------...--(((((....))))).................((((((((((.........))))))))))............((((-((.((......))...))))))----- ( -27.80, z-score = -3.05, R) >consensus ___CACCGACGCC__GGCUCUGAAGUGCCUUUUUCAAUUUCCCCAGAGGCGCUUCCGAUCAAACAGGAAGUGCUUAAGUUCAAACUGAAGUAAAAGUGCGCUCAGACCCA_CCCC____ .........(((...(((........)))..(((((.(((......((((((((((.........))))))))))......))).))))).......)))................... (-22.41 = -21.99 + -0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:43 2011