
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,825,539 – 23,825,645 |
| Length | 106 |
| Max. P | 0.873276 |

| Location | 23,825,539 – 23,825,641 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 86.98 |
| Shannon entropy | 0.26555 |
| G+C content | 0.36720 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -16.71 |
| Energy contribution | -16.62 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.873276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
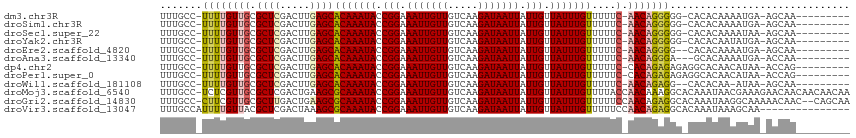
>dm3.chr3R 23825539 102 + 27905053 UUUGCC-UUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-AACAGGGGG-CACACAAAAUGA-AGCAA--------- ..((((-((((((((.((((.....))))(((((((.(((.(((((((.....))))))).))).)))))))....)-)))))))))-))..........-.....--------- ( -30.60, z-score = -3.12, R) >droSim1.chr3R 23527480 102 + 27517382 UUUGCC-UUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-AACAGGGGG-CACACAAAAUGA-AGCAA--------- ..((((-((((((((.((((.....))))(((((((.(((.(((((((.....))))))).))).)))))))....)-)))))))))-))..........-.....--------- ( -30.60, z-score = -3.12, R) >droSec1.super_22 448468 102 + 1066717 UUUGCC-UUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-AACAGGGGG-CACACAAAAUAA-AGCAA--------- ..((((-((((((((.((((.....))))(((((((.(((.(((((((.....))))))).))).)))))))....)-)))))))))-))..........-.....--------- ( -30.60, z-score = -3.46, R) >droYak2.chr3R 6158299 102 - 28832112 UUUGCC-UUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-AACAGGGGG-CACACAAUAUGA-AGCAA--------- ..((((-((((((((.((((.....))))(((((((.(((.(((((((.....))))))).))).)))))))....)-)))))))))-))..........-.....--------- ( -30.60, z-score = -3.25, R) >droEre2.scaffold_4820 6250688 101 - 10470090 UUUGCC-UUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-AACAGGGG--CACACAAAAUGA-AGCAA--------- ..((((-(((.((((.((((.....))))(((((((.(((.(((((((.....))))))).))).)))))))....)-))))))))--))..........-.....--------- ( -26.30, z-score = -1.94, R) >droAna3.scaffold_13340 21964033 100 + 23697760 UUUGCC-UUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-AACAGGGA---GCACAAAAUGA-ACCAA--------- ..(((.-.(((((((.((((.....))))(((((((.(((.(((((((.....))))))).))).)))))))....)-))))))..---)))........-.....--------- ( -24.20, z-score = -1.69, R) >dp4.chr2 22533399 103 - 30794189 UUUGCC-UUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-CACAGAGAGAGGCACAACAUAA-ACCAG--------- ..((((-((((.(((.((((.....))))(((((((.(((.(((((((.....))))))).))).))))))).....-..))).))))))))........-.....--------- ( -26.30, z-score = -2.71, R) >droPer1.super_0 9726506 103 - 11822988 UUUGCC-UUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-CACAGAGAGAGGCACAACAUAA-ACCAG--------- ..((((-((((.(((.((((.....))))(((((((.(((.(((((((.....))))))).))).))))))).....-..))).))))))))........-.....--------- ( -26.30, z-score = -2.71, R) >droWil1.scaffold_181108 1038058 100 + 4707319 UUUGCC-UUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-AACAGAGG--CACACAA-AUAA-AGCAA--------- ..((((-(((.((((.((((.....))))(((((((.(((.(((((((.....))))))).))).)))))))....)-))))))))--)).....-....-.....--------- ( -27.10, z-score = -2.85, R) >droMoj3.scaffold_6540 14888571 114 - 34148556 UUUGCC-UCUCGUUGCGCUCGACUGAAGCGCAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUACCAACAAAGGCACAAAUAACGAAAGAACAACAACAACAA ..((((-(....(((((((.......)))))))..........((((((..(((((((.((....)).)))))))..))))))))))).......(....).............. ( -26.30, z-score = -2.28, R) >droGri2.scaffold_14830 4257610 112 + 6267026 UUUGCC-CUUCGUUGCGCUUGACUGAAGCGCAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUCCAACAGAGGCACAAAUAAGGCAAAAACAAC--CAGCAA ((((((-.((((((((((((.....))))))))....))))..((((((..(((((((.((....)).)))))))..))))))............))))))......--...... ( -28.40, z-score = -1.91, R) >droVir3.scaffold_13047 3469978 100 - 19223366 UUUGCCAUUUUGUUACGCUCGACUAAAGCGCAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUCCAACAGAGGCACAAAUAAAGCAA--------------- ..((((..((((((..(((.......)))(((((((.(((.(((((((.....))))))).))).)))))))......))))))))))............--------------- ( -20.10, z-score = -1.06, R) >consensus UUUGCC_UUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC_AACAGAGGG_CACACAAAAUAA_AGCAA_________ .......(((((((..((((.....))))(((((((.(((.(((((((.....))))))).))).)))))))......))))))).............................. (-16.71 = -16.62 + -0.09)

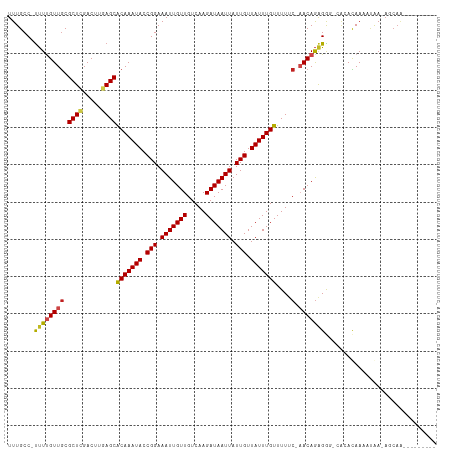
| Location | 23,825,543 – 23,825,645 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 86.71 |
| Shannon entropy | 0.27466 |
| G+C content | 0.37845 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.16 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.727045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23825543 102 + 27905053 -CCUUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-AACAGGGGG-CACACAAAAUGA-AGCAACAAC--------- -((((((((((.((((.....))))(((((((.(((.(((((((.....))))))).))).)))))))....)-)))))))))-............-.........--------- ( -25.30, z-score = -1.81, R) >droSim1.chr3R 23527484 102 + 27517382 -CCUUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-AACAGGGGG-CACACAAAAUGA-AGCAACAAC--------- -((((((((((.((((.....))))(((((((.(((.(((((((.....))))))).))).)))))))....)-)))))))))-............-.........--------- ( -25.30, z-score = -1.81, R) >droSec1.super_22 448472 102 + 1066717 -CCUUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-AACAGGGGG-CACACAAAAUAA-AGCAACAAC--------- -((((((((((.((((.....))))(((((((.(((.(((((((.....))))))).))).)))))))....)-)))))))))-............-.........--------- ( -25.30, z-score = -2.18, R) >droYak2.chr3R 6158303 102 - 28832112 -CCUUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-AACAGGGGG-CACACAAUAUGA-AGCAACAUC--------- -((((((((((.((((.....))))(((((((.(((.(((((((.....))))))).))).)))))))....)-)))))))))-............-.........--------- ( -25.30, z-score = -1.88, R) >droEre2.scaffold_4820 6250692 101 - 10470090 -CCUUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-AACAGGGG--CACACAAAAUGA-AGCAACAAC--------- -....(((((((((((...(((((.(((((((.(((.(((((((.....))))))).))).)))))))..)))-))...)))--).((.....)).-.))))))).--------- ( -25.30, z-score = -2.06, R) >droAna3.scaffold_13340 21964037 100 + 23697760 -CCUUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-AACAGGGA---GCACAAAAUGA-ACCAACAAC--------- -..((((((.((.(((...(((((.(((((((.(((.(((((((.....))))))).))).)))))))..)))-))..))).---))))))))...-.........--------- ( -22.40, z-score = -1.45, R) >dp4.chr2 22533403 103 - 30794189 -CCUUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-CACAGAGAGAGGCACAACAUAA-ACCAGCAAC--------- -......(((((((((.....)))).........((....((((((..(((((((....)))))))(((((((-......)))))))))))))...-.)).)))))--------- ( -26.60, z-score = -2.86, R) >droPer1.super_0 9726510 103 - 11822988 -CCUUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-CACAGAGAGAGGCACAACAUAA-ACCAGCAAC--------- -......(((((((((.....)))).........((....((((((..(((((((....)))))))(((((((-......)))))))))))))...-.)).)))))--------- ( -26.60, z-score = -2.86, R) >droWil1.scaffold_181108 1038062 100 + 4707319 -CCUUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC-AACAGAGG--CACACAA-AUAA-AGCAAGAAC--------- -.(((((((((.((((.....))))(((((((.(((.(((((((.....))))))).))).)))))))....)-))))))))--.......-....-.........--------- ( -23.20, z-score = -1.88, R) >droMoj3.scaffold_6540 14888575 114 - 34148556 -CCUCUCGUUGCGCUCGACUGAAGCGCAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUACCAACAAAGGCACAAAUAACGAAAGAACAACAACAACAACAGC -((.....(((((((.......))))))).....))...(((((((...........(((((((((((....((......)).)))))))))))..........))))))).... ( -26.10, z-score = -2.46, R) >droGri2.scaffold_14830 4257614 112 + 6267026 -CCCUUCGUUGCGCUUGACUGAAGCGCAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUCCAACAGAGGCACAAAUAAGGCAAAAACAAC--CAGCAACAGU -((.....((((((((.....)))))))).....))..((((((((..............((((((((.((((.....)))).))))))))((.........)--).)))))))) ( -26.50, z-score = -1.61, R) >droVir3.scaffold_13047 3469982 100 - 19223366 CCAUUUUGUUACGCUCGACUAAAGCGCAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUCCAACAGAGG----CACAA-AUAA-AGCAACAGC--------- ((..((((...((((.......))))))))....))...((((((((((........)))((((((((.((((.....))))----.))))-))))-.))))))).--------- ( -16.60, z-score = -0.07, R) >consensus _CCUUUUGUUGCGCUCGACUUGAGCACAAAUACCGGAAAUUGUUGUCAAGAUAAUUAUUGUUAUUUGUUUUUC_AACAGAGGG_CACACAAAAUAA_AGCAACAAC_________ .....((((((.((((.....))))(((((((.(((.(((((((.....))))))).))).)))))))...............................)))))).......... (-17.00 = -17.16 + 0.15)


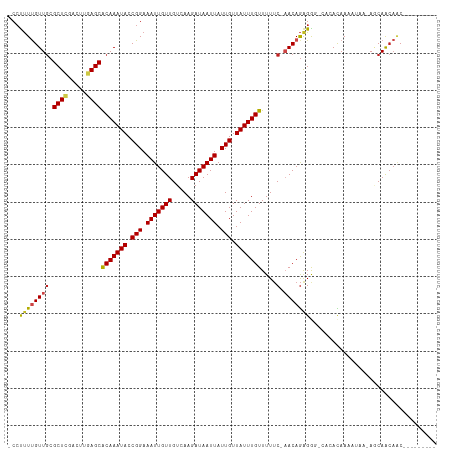
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:42 2011