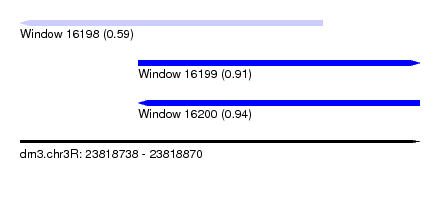
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,818,738 – 23,818,870 |
| Length | 132 |
| Max. P | 0.938243 |

| Location | 23,818,738 – 23,818,838 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 89.15 |
| Shannon entropy | 0.22657 |
| G+C content | 0.42707 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -21.78 |
| Energy contribution | -21.71 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.587247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
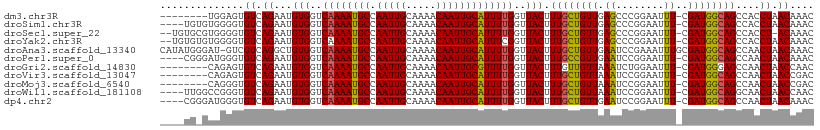
>dm3.chr3R 23818738 100 - 27905053 --------UGGAGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUUGGUUACUUUGCUGUUGAGCCCGGAAUUU-CGAUGGCAGCCACCUAACAAAC --------(((.(((.(.....(((..((((((((.(((((.....)))))))))))))..)))..((((((((((........))-))))))))).))))))...... ( -30.30, z-score = -1.89, R) >droSim1.chr3R 23520754 104 - 27517382 ----UGUGUGGGGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUUGGUUACUUUGCUGUUGAGCCCGGAAUUU-CGAUGGCAGCCACCUAACAAAC ----..(((.(((((.(.....(((..((((((((.(((((.....)))))))))))))..)))..((((((((((........))-))))))))).))))).)))... ( -35.90, z-score = -2.54, R) >droSec1.super_22 441707 105 - 1066717 --UGUGCGUGGGGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUUGGUUACUUUGCUGUUGAGCCCGGAAUUU-CGAUGGCAGCCACCU-ACAAAC --((((.((((...........(((..((((((((.(((((.....)))))))))))))..))).(((((((((((........))-))))))))))))).)-)))... ( -35.50, z-score = -2.02, R) >droYak2.chr3R 6151321 106 + 28832112 --UGUGUGUGGGGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUCGGUUACUUUGCUGUUGAGCCCGGAAUUU-CGAUGGCAGCCACCUAACAAAC --....(((.(((((.(.....(((..(.((((((.(((((.....))))))))))).)..)))..((((((((((........))-))))))))).))))).)))... ( -31.80, z-score = -1.04, R) >droAna3.scaffold_13340 21957156 108 - 23697760 CAUAUGGGAU-GUCGUCAGGCUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUUGGUUACUUUGCUGUUGAAUCCGAAAUUUGCGAUGGCAGCCAACUAACAAAC ....(((..(-((((((.(((.(((..((((((((.(((((.....)))))))))))))..)))...))).(((....))).......))))))).))).......... ( -32.40, z-score = -2.06, R) >droPer1.super_0 9719040 104 + 11822988 ----CGGGAUGGGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUUGGUUACUUUGCCGUUGAAUCCGGAAUUU-CGAUGGCAGCCAACUAACAAAC ----..((.(((..........(((..((((((((.(((((.....)))))))))))))..))).(((((((((((........))-)))))))))))).))....... ( -34.50, z-score = -2.84, R) >droGri2.scaffold_14830 4249585 100 - 6267026 --------CAGAGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCGUUUUGGUUACUUUGUUGUUAAAUCUGGAAUUU-CGAUGGGAGCCAACUAACCAAC --------((((....(((...(((..((((((((.(((((.....)))))))))))))..)))....))).....))))......-...(((...........))).. ( -24.10, z-score = -0.90, R) >droVir3.scaffold_13047 3459981 100 + 19223366 --------CAGAGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUUGGUUACUUUGCUGUUAAAUCCGGAAUUU-CGAUGGCAGCCAACUAACCGAC --------......(((.....(((..((((((((.(((((.....)))))))))))))..)))...(((((((....(((....)-)).))))))).........))) ( -25.70, z-score = -1.33, R) >droMoj3.scaffold_6540 14877277 100 + 34148556 --------CAGGGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUUGGUUACUUUGCUGUUAAAUCCGGAAUUU-CGAUGGCAGCCAACUAACCGAC --------..((.(((((....(((..((((((((.(((((.....)))))))))))))..)))..............(((....)-)).))))).))........... ( -27.10, z-score = -1.35, R) >droWil1.scaffold_181108 1029628 104 - 4707319 ----UUGGCCGGGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUUGGUUACUUUGCUGUUAAAUCCGGAAUUU-CGAUGGCAGGCAACUAACCAAC ----((((((((((..(((...(((..((((((((.(((((.....)))))))))))))..)))....)))....)))))).....-.......((....))..)))). ( -34.60, z-score = -2.91, R) >dp4.chr2 22525941 104 + 30794189 ----CGGGAUGGGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUUGGUUACUUUGCUGUUGAAUCCGGAAUUU-CGAUGGCAGCCAACUAACAAAC ----..((.(((.((((((...(((..((((((((.(((((.....)))))))))))))..)))....))((((((........))-)))))))).))).))....... ( -31.70, z-score = -2.25, R) >consensus ____UG_GCGGGGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUUGGUUACUUUGCUGUUGAAUCCGGAAUUU_CGAUGGCAGCCAACUAACAAAC ..............((.((...(((..((((((((.(((((.....)))))))))))))..))).((((((((...............))))))))....)).)).... (-21.78 = -21.71 + -0.06)
| Location | 23,818,777 – 23,818,870 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 74.57 |
| Shannon entropy | 0.46935 |
| G+C content | 0.41623 |
| Mean single sequence MFE | -17.95 |
| Consensus MFE | -12.25 |
| Energy contribution | -12.26 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.910993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23818777 93 + 27905053 GCAAAGUAACCAAAAUGCAAUUGUUUUGCAAUUGGCAUUUUGACCACAUUCUGACACUCCAC------ACACAGAUACAAACACCAAUACAUAUGGACG .....(((..(((((((((((((.....))))).)))))))).......((((.........------...))))))).....(((.......)))... ( -16.80, z-score = -1.91, R) >droSim1.chr3R 23520793 97 + 27517382 GCAAAGUAACCAAAAUGCAAUUGUUUUGCAAUUGGCAUUUUGACCACAUUCUGACACCCCAC--ACACACACAGAUACAAACACCAAUACAUAUGGCCG .....(((..(((((((((((((.....))))).)))))))).......((((.........--.......))))))).....(((.......)))... ( -15.79, z-score = -1.13, R) >droSec1.super_22 441745 99 + 1066717 GCAAAGUAACCAAAAUGCAAUUGUUUUGCAAUUGGCAUUUUGACCACAUUCUGACACCCCACGCACACACACAGAUACAAACACCAAUACAUAUGGGCG ((...(((..(((((((((((((.....))))).)))))))).......((((..................))))))).....(((.......))))). ( -18.77, z-score = -1.37, R) >droYak2.chr3R 6151360 99 - 28832112 GCAAAGUAACCGAAAUGCAAUUGUUUUGCAAUUGGCAUUUUGACCACAUUCUGACACCCCACACACACACACAGAUACAAGCACUAAUACAUACGGACG ((........(((((((((((((.....))))).)))))))).......((((..................)))).....))................. ( -15.67, z-score = -1.46, R) >droGri2.scaffold_14830 4249624 73 + 6267026 ACAAAGUAACCAAAACGCAAUUGUUUUGCAAUUGGCAUUUUGACCACAUUCUGACACUCUGG--CAAUAUGGCCA------------------------ .((((((..((((...((((.....))))..)))).))))))..................((--(......))).------------------------ ( -14.00, z-score = -0.32, R) >droVir3.scaffold_13047 3460020 73 - 19223366 GCAAAGUAACCAAAAUGCAAUUGUUUUGCAAUUGGCAUUUUGACCACAUUCUGACACUCUGG--CCAUAUGGCCA------------------------ ..........(((((((((((((.....))))).))))))))..................((--((....)))).------------------------ ( -20.60, z-score = -2.03, R) >droMoj3.scaffold_6540 14877316 73 - 34148556 GCAAAGUAACCAAAAUGCAAUUGUUUUGCAAUUGGCAUUUUGACCACAUUCUGACACCCUGG--CCAUAUGGCCG------------------------ ..........(((((((((((((.....))))).))))))))..................((--((....)))).------------------------ ( -20.60, z-score = -2.14, R) >droWil1.scaffold_181108 1029667 85 + 4707319 GCAAAGUAACCAAAAUGCAAUUGUUUUGCAAUUGGCAUUUUGACCACAUUCUGACAC-CCGG--CCA-AUGGCAAUGCCAAUACGGCUA---------- ..........(((((((((((((.....))))).))))))))...............-..((--((.-.((((...))))....)))).---------- ( -21.40, z-score = -0.72, R) >consensus GCAAAGUAACCAAAAUGCAAUUGUUUUGCAAUUGGCAUUUUGACCACAUUCUGACACCCCAC__CCACACACAGAUACAAACAC_AAUA__________ ..........(((((((((((((.....))))).))))))))......................................................... (-12.25 = -12.26 + 0.02)

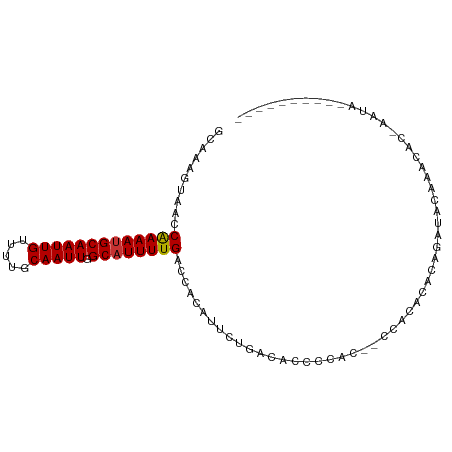
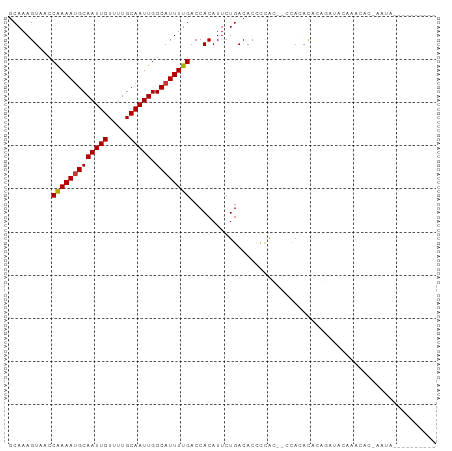
| Location | 23,818,777 – 23,818,870 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 74.57 |
| Shannon entropy | 0.46935 |
| G+C content | 0.41623 |
| Mean single sequence MFE | -25.29 |
| Consensus MFE | -16.28 |
| Energy contribution | -16.30 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
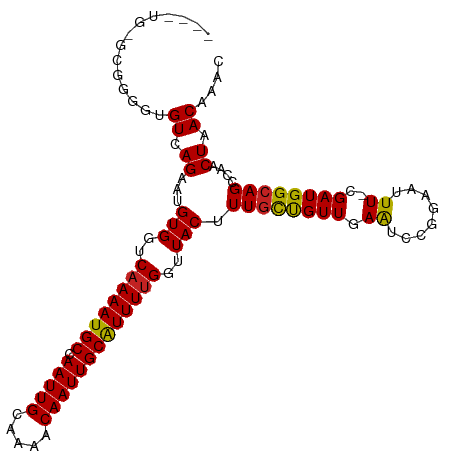
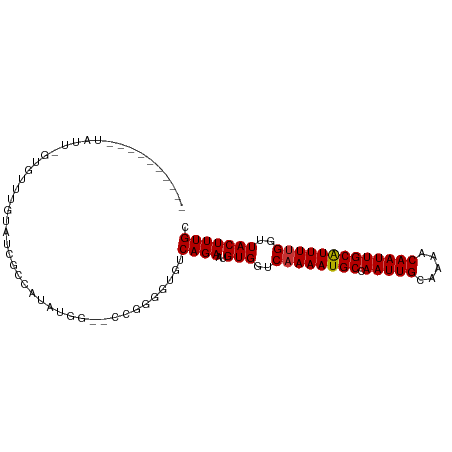
>dm3.chr3R 23818777 93 - 27905053 CGUCCAUAUGUAUUGGUGUUUGUAUCUGUGU------GUGGAGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUUGGUUACUUUGC ..((((((..((..((((....))))))..)------)))))..........(((..((((((((.(((((.....)))))))))))))..)))..... ( -25.90, z-score = -2.03, R) >droSim1.chr3R 23520793 97 - 27517382 CGGCCAUAUGUAUUGGUGUUUGUAUCUGUGUGUGU--GUGGGGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUUGGUUACUUUGC ...(((((..(((.((((....))))...)))..)--))))...........(((..((((((((.(((((.....)))))))))))))..)))..... ( -27.20, z-score = -1.39, R) >droSec1.super_22 441745 99 - 1066717 CGCCCAUAUGUAUUGGUGUUUGUAUCUGUGUGUGUGCGUGGGGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUUGGUUACUUUGC (((((.(((((((..(..(........)..)..)))))))))))).......(((..((((((((.(((((.....)))))))))))))..)))..... ( -29.80, z-score = -1.70, R) >droYak2.chr3R 6151360 99 + 28832112 CGUCCGUAUGUAUUAGUGCUUGUAUCUGUGUGUGUGUGUGGGGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUCGGUUACUUUGC ...((.((..(((..(..(........)..)..)))..)).)).........(((..(.((((((.(((((.....))))))))))).)..)))..... ( -20.30, z-score = 0.72, R) >droGri2.scaffold_14830 4249624 73 - 6267026 ------------------------UGGCCAUAUUG--CCAGAGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCGUUUUGGUUACUUUGU ------------------------((((......)--)))......((((..(((..((((((((.(((((.....)))))))))))))..))))))). ( -21.00, z-score = -1.69, R) >droVir3.scaffold_13047 3460020 73 + 19223366 ------------------------UGGCCAUAUGG--CCAGAGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUUGGUUACUUUGC ------------------------(((((....))--)))............(((..((((((((.(((((.....)))))))))))))..)))..... ( -25.30, z-score = -2.78, R) >droMoj3.scaffold_6540 14877316 73 + 34148556 ------------------------CGGCCAUAUGG--CCAGGGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUUGGUUACUUUGC ------------------------.((((....))--)).............(((..((((((((.(((((.....)))))))))))))..)))..... ( -24.90, z-score = -2.34, R) >droWil1.scaffold_181108 1029667 85 - 4707319 ----------UAGCCGUAUUGGCAUUGCCAU-UGG--CCGG-GUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUUGGUUACUUUGC ----------((((((.(((((((((....(-(((--(((.-((......)).))))))))))))))))(((((.....)))))...))))))...... ( -27.90, z-score = -1.64, R) >consensus __________UAUU_GUGUUUGUAUCGCCAUAUGG__CCGGGGUGUCAGAAUGUGGUCAAAAUGCCAAUUGCAAAACAAUUGCAUUUUGGUUACUUUGC ..............................................((((..(((..((((((((.(((((.....)))))))))))))..))))))). (-16.28 = -16.30 + 0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:38 2011