
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,807,240 – 23,807,302 |
| Length | 62 |
| Max. P | 0.995349 |

| Location | 23,807,240 – 23,807,302 |
|---|---|
| Length | 62 |
| Sequences | 6 |
| Columns | 77 |
| Reading direction | forward |
| Mean pairwise identity | 70.74 |
| Shannon entropy | 0.50236 |
| G+C content | 0.36410 |
| Mean single sequence MFE | -13.42 |
| Consensus MFE | -6.99 |
| Energy contribution | -7.05 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.903023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
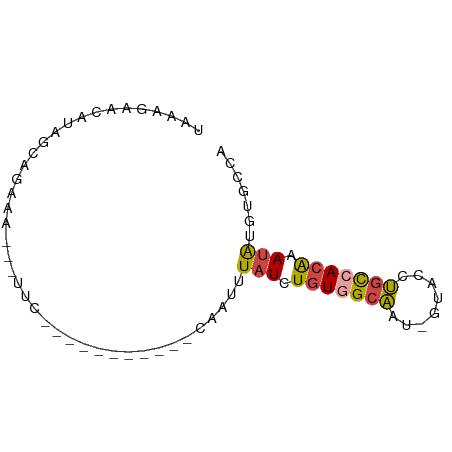
>dm3.chr3R 23807240 62 + 27905053 UAAAGAACAUAGCUGAAA---CUC-----------CAAUUUAUCUGUAGCAAU-GUACCUGUCACAAAUAUGUGCCA ......((((.((((...---...-----------...........)))).))-))....(.((((....)))).). ( -6.93, z-score = 0.11, R) >droEre2.scaffold_4820 6232347 76 - 10470090 UAAAGAACAUAGCAGAAACGGUCCAAAAAUUUAUCCAAUUUAUCUGUGGCAAC-GUAGCUGCCACAAAUAUCUGCCG ...........(((((...((.............))....(((.(((((((..-.....))))))).)))))))).. ( -17.82, z-score = -3.06, R) >droYak2.chr3R 6139666 74 - 28832112 UAAAGAACAUGGCAGAAACGGUUCAAAAACUU---CAAUUUAUCUGUGGCAACUGUGGCUGCCACAAAUAUCUGCCG ..........((((((...((((....)))).---.....(((.(((((((.(....).))))))).))))))))). ( -22.00, z-score = -3.59, R) >droSec1.super_22 429650 62 + 1066717 UAAAGAACAUAGCGGAAA---UUC-----------CAAUUUAUCUGUGGCAAU-GUACCUGCCACAAAUAUGUCCCA ......(((((..((...---..)-----------)........(((((((..-.....)))))))..))))).... ( -14.60, z-score = -2.51, R) >droSim1.chr3R 23509335 62 + 27517382 UAAAGAACAUAGCAGAAA---UUC-----------CAAUUUAUCUGUGGCAAU-GUACCUGCCACAAAUAUGUGCCA ......((((((((((.(---...-----------.....).)))))((((..-.....)))).....))))).... ( -11.70, z-score = -1.20, R) >droVir3.scaffold_13047 6833566 62 + 19223366 UAAAGAACAAGGCUGAUA---UUCA-----------UAUUUGUUUUU-GUUUUGUUACCGUUCAUGUAAGUUUAUAA .....((((((((.((((---....-----------....))))...-))))))))..................... ( -7.50, z-score = -0.18, R) >consensus UAAAGAACAUAGCAGAAA___UUC___________CAAUUUAUCUGUGGCAAU_GUACCUGCCACAAAUAUGUGCCA ........................................(((.(((((((........))))))).)))....... ( -6.99 = -7.05 + 0.06)

| Location | 23,807,240 – 23,807,302 |
|---|---|
| Length | 62 |
| Sequences | 6 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 70.74 |
| Shannon entropy | 0.50236 |
| G+C content | 0.36410 |
| Mean single sequence MFE | -18.05 |
| Consensus MFE | -9.69 |
| Energy contribution | -11.42 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.995349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
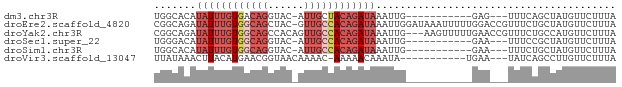
>dm3.chr3R 23807240 62 - 27905053 UGGCACAUAUUUGUGACAGGUAC-AUUGCUACAGAUAAAUUG-----------GAG---UUUCAGCUAUGUUCUUUA ((((...((((((((.(((....-.))).))))))))...((-----------(..---..)))))))......... ( -9.80, z-score = 0.39, R) >droEre2.scaffold_4820 6232347 76 + 10470090 CGGCAGAUAUUUGUGGCAGCUAC-GUUGCCACAGAUAAAUUGGAUAAAUUUUUGGACCGUUUCUGCUAUGUUCUUUA .(((((((((((((((((((...-)))))))))))))..(..((......))..)......)))))).......... ( -26.30, z-score = -4.28, R) >droYak2.chr3R 6139666 74 + 28832112 CGGCAGAUAUUUGUGGCAGCCACAGUUGCCACAGAUAAAUUG---AAGUUUUUGAACCGUUUCUGCCAUGUUCUUUA .(((((((((((((((((((....))))))))))))).....---..(((....)))....)))))).......... ( -28.60, z-score = -5.33, R) >droSec1.super_22 429650 62 - 1066717 UGGGACAUAUUUGUGGCAGGUAC-AUUGCCACAGAUAAAUUG-----------GAA---UUUCCGCUAUGUUCUUUA .((((((((((((((((((....-.))))))))))))....(-----------(..---...))....))))))... ( -22.30, z-score = -4.20, R) >droSim1.chr3R 23509335 62 - 27517382 UGGCACAUAUUUGUGGCAGGUAC-AUUGCCACAGAUAAAUUG-----------GAA---UUUCUGCUAUGUUCUUUA ((((...((((((((((((....-.))))))))))))....(-----------(..---...))))))......... ( -18.70, z-score = -2.78, R) >droVir3.scaffold_13047 6833566 62 - 19223366 UUAUAAACUUACAUGAACGGUAACAAAAC-AAAAACAAAUA-----------UGAA---UAUCAGCCUUGUUCUUUA ..............(((((((........-...........-----------....---.....)))..)))).... ( -2.59, z-score = 0.70, R) >consensus UGGCACAUAUUUGUGGCAGGUAC_AUUGCCACAGAUAAAUUG___________GAA___UUUCUGCUAUGUUCUUUA .......((((((((((((......))))))))))))........................................ ( -9.69 = -11.42 + 1.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:35 2011