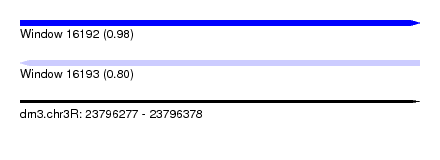
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,796,277 – 23,796,378 |
| Length | 101 |
| Max. P | 0.979105 |

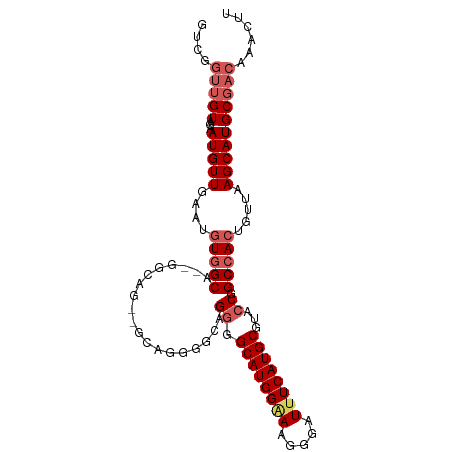
| Location | 23,796,277 – 23,796,378 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 79.98 |
| Shannon entropy | 0.35665 |
| G+C content | 0.54156 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -20.14 |
| Energy contribution | -21.62 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.979105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23796277 101 + 27905053 GUCGGUUGUAGAAUGUUGAAUGUGGCACGGGCAGGGGCAGGGGCAGGGGCAUGGAAAGGGAUUUCAUGCGUACCGGCCACUGUUAAGCAUGCGACAAACUU ((..(((((...(((((.((((((((.(((((....((....))....((((((((.....)))))))))).)))))))).))).))))))))))..)).. ( -34.00, z-score = -1.88, R) >droEre2.scaffold_4820 6221246 76 - 10470090 -------GUCGGAUGUUGAAUGUGGCA------------------GGGGCAUGGUAAGGGAUUUCAUGCGGACCGGCCACUGCGAAGCAUGCGACAAACUU -------((((.(((((....(((((.------------------((.((((((.........))))))...)).))))).....))))).))))...... ( -31.40, z-score = -3.48, R) >droYak2.chr3R 6128323 86 - 28832112 GUCGGUUGUUGAAUGUUG----UUGCA-----------UGUGGCAGGGGCAUGGGAAGGGAUUUCAUGCCGAACGGCCACUGUUAAGCAUGCGACAAACUU .....((((((.(((((.----..(((-----------.(((((.(.((((((..(.....)..))))))...).))))))))..))))).)))))).... ( -33.80, z-score = -3.09, R) >droSec1.super_22 418745 101 + 1066717 GUCGGUUGUAGCAUGUUGAAUGUGGCAGGGGCAGGUGCUGGGGCAGGGGCAUGGAAAGGGAUUUCAUGCGUACCGGCCACUGUUAAGCAUGCGACAAACUU .....((((.(((((((.((((((((.((..(...(((....)))...((((((((.....)))))))))..)).))))).))).))))))).)))).... ( -38.80, z-score = -2.60, R) >droSim1.chr3R 23498385 101 + 27517382 GUCGGUUGUAGAAUGUUGAAUGUGGCAGUGGCAGUGGCAGGGGCAGGGGCAUGGAAAGGGAUUUCAUGCGUACCGGCCACUGUUAAGCAUGCGACAAACUU ((((..(((....((((......)))).((((((((((..((....(.((((((((.....)))))))).).)).)))))))))).)))..))))...... ( -37.00, z-score = -2.62, R) >consensus GUCGGUUGUAGAAUGUUGAAUGUGGCA__GGCAG__GCAGGGGCAGGGGCAUGGAAAGGGAUUUCAUGCGUACCGGCCACUGUUAAGCAUGCGACAAACUU ....(((((...(((((....(((((....((....)).......((.((((((((.....))))))))...)).))))).....))))))))))...... (-20.14 = -21.62 + 1.48)


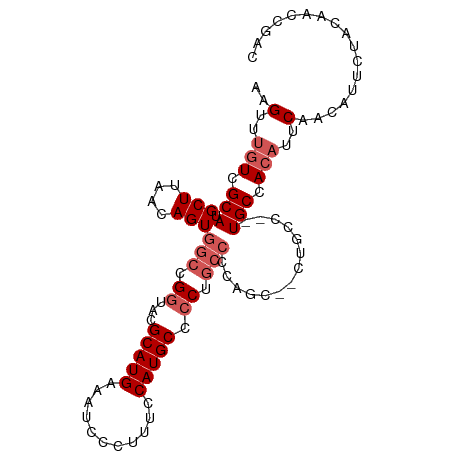
| Location | 23,796,277 – 23,796,378 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 79.98 |
| Shannon entropy | 0.35665 |
| G+C content | 0.54156 |
| Mean single sequence MFE | -23.91 |
| Consensus MFE | -11.84 |
| Energy contribution | -13.04 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.803780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23796277 101 - 27905053 AAGUUUGUCGCAUGCUUAACAGUGGCCGGUACGCAUGAAAUCCCUUUCCAUGCCCCUGCCCCUGCCCCUGCCCGUGCCACAUUCAACAUUCUACAACCGAC ..(..(((.(((((.....(((.(((.((((.(((((.((.....)).)))))...))))...))).)))..))))).)))..)................. ( -24.80, z-score = -2.37, R) >droEre2.scaffold_4820 6221246 76 + 10470090 AAGUUUGUCGCAUGCUUCGCAGUGGCCGGUCCGCAUGAAAUCCCUUACCAUGCCCC------------------UGCCACAUUCAACAUCCGAC------- ......((((.(((.......(((((.((...(((((...........))))).))------------------.)))))......))).))))------- ( -20.62, z-score = -2.02, R) >droYak2.chr3R 6128323 86 + 28832112 AAGUUUGUCGCAUGCUUAACAGUGGCCGUUCGGCAUGAAAUCCCUUCCCAUGCCCCUGCCACA-----------UGCAA----CAACAUUCAACAACCGAC ..(((.((.(((((.....).(((((.(...((((((...........)))))).).))))))-----------))).)----)))).............. ( -20.80, z-score = -1.74, R) >droSec1.super_22 418745 101 - 1066717 AAGUUUGUCGCAUGCUUAACAGUGGCCGGUACGCAUGAAAUCCCUUUCCAUGCCCCUGCCCCAGCACCUGCCCCUGCCACAUUCAACAUGCUACAACCGAC ..(.((((.(((((.......(((((.((..((((((.((.....)).)))))...(((....)))...)..)).)))))......))))).)))).)... ( -27.42, z-score = -2.82, R) >droSim1.chr3R 23498385 101 - 27517382 AAGUUUGUCGCAUGCUUAACAGUGGCCGGUACGCAUGAAAUCCCUUUCCAUGCCCCUGCCCCUGCCACUGCCACUGCCACAUUCAACAUUCUACAACCGAC ..(..(((.(((((.....(((((((.((((.(((((.((.....)).)))))...))))...))))))).)).))).)))..)................. ( -25.90, z-score = -2.82, R) >consensus AAGUUUGUCGCAUGCUUAACAGUGGCCGGUACGCAUGAAAUCCCUUUCCAUGCCCCUGCCCCAGC__CUGCC__UGCCACAUUCAACAUUCUACAACCGAC ..(..(((.(((.(((....)))(((.((...(((((...........))))).)).)))..............))).)))..)................. (-11.84 = -13.04 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:33 2011