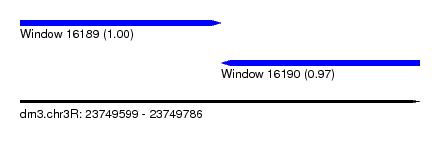
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,749,599 – 23,749,786 |
| Length | 187 |
| Max. P | 0.999735 |

| Location | 23,749,599 – 23,749,693 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 55.89 |
| Shannon entropy | 0.79314 |
| G+C content | 0.43791 |
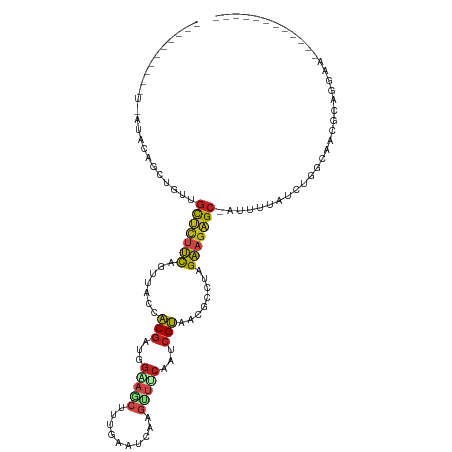
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -13.04 |
| Energy contribution | -12.26 |
| Covariance contribution | -0.79 |
| Combinations/Pair | 1.71 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.28 |
| SVM RNA-class probability | 0.999735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
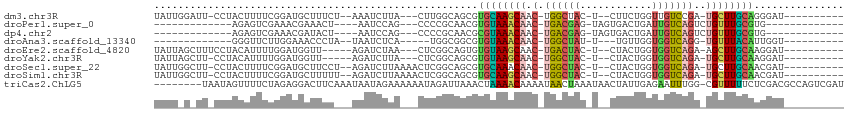
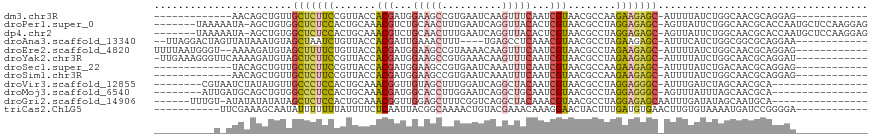
>dm3.chr3R 23749599 94 + 27905053 UAUUGGAUU-CCUACUUUUCGGAUGCUUUCU--AAAUCUUA---CUUGGCAGCGUGCAAGCAAC-UGGCUAC-U--CUUCUGGUUGUCCGA-UGCUUGCAGGGAU---------- ......(((-(((.........(((((..((--((......---.)))).)))))(((((((.(-.(((.((-(--.....))).))).).-)))))))))))))---------- ( -25.40, z-score = -1.14, R) >droPer1.super_0 9648785 80 - 11822988 -------------AGAGUCGAAACGAAACU----AAUCCAG---CCCCGCAACGUGUAAACAAC-UGACGAG-UAGUGACUGAUUGUCAGUCUGUUUGCGUG------------- -------------((..(((...)))..))----.......---........((((((((((((-((((((.-(((...))).)))))))).))))))))))------------- ( -18.60, z-score = -0.85, R) >dp4.chr2 22455081 80 - 30794189 -------------AGAGUCGAAACGAUACU----AAUCCAG---CCCCGCAACGCGUAAACAAC-UGACGAG-UAGUGACUGAUUGUCAGUCUGUUUGCGUG------------- -------------((.((((...)))).))----.......---........((((((((((((-((((((.-(((...))).)))))))).))))))))))------------- ( -24.60, z-score = -2.72, R) >droAna3.scaffold_13340 21885567 79 + 23697760 -------------GGGUUCUUGGAAACCCUA--UAAUCUCA-----UGGCGGCGUGUAAACAAC-UGGCUAU-U---UGUUGGUGGUCAGG-UGUUUACAUUGGU---------- -------------(((((......)))))..--........-----..((.(.(((((((((.(-(((((((-(---....))))))))).-)))))))))).))---------- ( -26.20, z-score = -2.76, R) >droEre2.scaffold_4820 6177161 92 - 10470090 UAUUAGCUUUCCUACAUUUUGGAUGGUU-----AGAUCUAA---CUCGGCAGUGUGUAAGCAAC-UGACUAC-U--CUACUGGUGGUCAGA-AGCUUGCAAGGAU---------- ..((((((.(((........))).))))-----))((((.(---((....))).(((((((..(-(((((((-(--.....))))))))).-.))))))).))))---------- ( -31.40, z-score = -3.05, R) >droYak2.chr3R 6083982 91 - 28832112 UAUUAGCUU-CCUACAUUUUGGAUGGUU-----AGAUCUUA---CUCGGCAGCGUGUAAGCAAC-UGGCUAC-U--CUACUGGUGGUCAGA-UGCUUGCAAGGAU---------- ..(((((((-((........))).))))-----))(((((.---..((....))((((((((.(-(((((((-(--.....))))))))).-)))))))))))))---------- ( -31.00, z-score = -2.58, R) >droSec1.super_22 375030 97 + 1066717 UAUUGGCUU-CCUACUUUUCGGAUGCUUCCU--AGAUCUUAAAACUCGGCAGCGUGCAAACAAC-UGGCUAC-U--CUACUGGUGGUCAGA-UGCUUGCAACGAU---------- .(((((((.-((...((((.((((.((....--)))))).))))...)).))).(((((.((.(-(((((((-(--.....))))))))).-)).))))).))))---------- ( -28.80, z-score = -2.69, R) >droSim1.chr3R 23459619 97 + 27517382 UAUUGGCUU-CCUACUUUUCGGAUGCUUUUU--AGAUCUUAAAACUCGGCAGCGUGCAAGCAAC-UGGCUAC-U--CUACUGGUGGUCAGA-UGCUUGCAACGAU---------- .(((((((.-((...((((.((((.((....--)))))).))))...)).))).((((((((.(-(((((((-(--.....))))))))).-)))))))).))))---------- ( -35.40, z-score = -4.40, R) >triCas2.ChLG5 2832769 106 - 18847211 --------UAAUAGUUUUCUAGAGGACUUCAAAUAAUAGAAAAAAUAGAUUAAACUAAAACAAAAUAACUAAAUAACUAUUGAGAAUUUGG-CGUUUUUCUCGACGCCAGUCGAU --------((((((((...(((.....(((........)))....(((......)))...........)))...)))))))).((...(((-((((......))))))).))... ( -17.50, z-score = -1.52, R) >consensus UAUU_G_UU_CCUACAUUUUGGAUGAUUCU___AAAUCUAA___CUCGGCAGCGUGUAAACAAC_UGGCUAC_U__CUACUGGUGGUCAGA_UGCUUGCAAGGAU__________ ......................................................((((((((...((((((............))))))...))))))))............... (-13.04 = -12.26 + -0.79)
| Location | 23,749,693 – 23,749,786 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 60.33 |
| Shannon entropy | 0.79218 |
| G+C content | 0.44537 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -11.57 |
| Energy contribution | -11.53 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.974828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
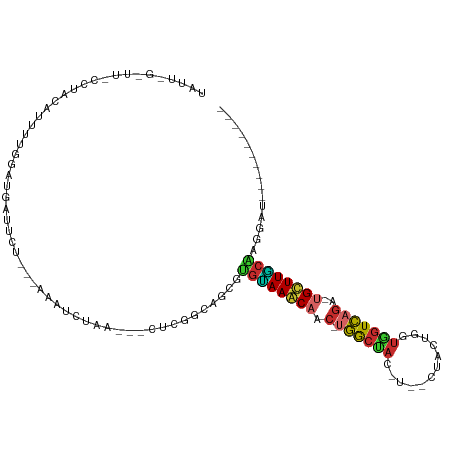
>dm3.chr3R 23749693 93 - 27905053 -------------AACAGCUGUUGCUCUUCCGUUACCACGAUGGAAGCCGUGAAUCAAGUUUCAAUCGUAACGCCAAGAAGAGC-AUUUUAUCUGGCAACGCAGGAG------------ -------------.....((((((((((((((((...(((((.(((((..........))))).)))))))))....)))))))-)........(....)))))...------------ ( -25.80, z-score = -1.63, R) >droPer1.super_0 9648865 110 + 11822988 -------UAAAAAUA-AGCUGUGGCUCUCCACUGCAAACGUCUGCAACUUUGAAUCAGGUUACACUCGUAACGCCUAGGAGAGC-AGUUAUUCUGGCAACGCACCAAUGCUCCAAGGAG -------........-.(.(((((((((((..((((......))))............(((((....))))).....)))))))-.(((.....)))..)))).)....(((....))) ( -27.80, z-score = -0.64, R) >dp4.chr2 22455161 110 + 30794189 -------UAAAAAUA-AGCUGUGGCUCUCCACUGCAAACGUCUGCAACUUUGAAUCAGGUUACACUCGUAACGCCUAGGAGAGC-AGUUAUUCUGGCAACGCACCAAUGCUCCAAGGAG -------........-.(.(((((((((((..((((......))))............(((((....))))).....)))))))-.(((.....)))..)))).)....(((....))) ( -27.80, z-score = -0.64, R) >droAna3.scaffold_13340 21885646 100 - 23697760 --UUAGGACUAGUUAUAAAUGUAGCUAAUCUGUUACCACGAUUGAAACUUU----UGAGCCUCAAACGUAACGCCUAGAAGAGC-AUUUCAUCUGGCGGCGCAGGAA------------ --...(((.(((((((....))))))).)))....(((((.((((..((..----..))..)))).)))..((((.(((.(((.-..))).))))))).....))..------------ ( -23.70, z-score = -0.77, R) >droEre2.scaffold_4820 6177253 104 + 10470090 UUUUAAUGGGU--AAAAGAUGUAGCUUUUCUGUUACCACGAUGGAAGCCGUAAAACAAGUUUCAAUCGUAACGCCUAGAAGAGC-AUUUUAUCUGGCAACGCAGGAG------------ ......((.((--...(((((..(((((((((.....(((((.(((((..........))))).)))))......)))))))))-....)))))....)).))....------------ ( -24.40, z-score = -1.02, R) >droYak2.chr3R 6084073 105 + 28832112 -UUGAAAGGGUUCAAAAGAUGUAGCUCUUCCGUUACCACGAUGGAAGCCGUGAAACAAGUUUCAAUCGUAACGCCUAGAAGAGC-AUUUUAUCUGGCAACGCAGGAU------------ -......(.((((...(((((..(((((((((((...(((((.(((((..........))))).)))))))))....)))))))-....))))).).))).).....------------ ( -26.60, z-score = -1.11, R) >droSec1.super_22 375127 93 - 1066717 -------------UACAGCUGUUGCUCUUCCGUUACCACGAUGGAAGCCGUGAAUCAAAUUUCAAUCGUAACGCCAAGAAGAGC-AUUUUAUCUGACAACGCAGGAG------------ -------------..(((.((.((((((((((((((....((((...))))(((......)))....))))))....)))))))-)...)).)))............------------ ( -22.00, z-score = -1.24, R) >droSim1.chr3R 23459716 93 - 27517382 -------------AACAGCUGUUGCUCUUCCGUUACCACGAUGGAAGCCGUGAAUCAAAUUUCAAUCGUAACGCCAAGAAGAGC-AUUUUAUCUGGCAACGCAGGAG------------ -------------.....((((((((((((((((((....((((...))))(((......)))....))))))....)))))))-)........(....)))))...------------ ( -23.80, z-score = -1.29, R) >droVir3.scaffold_12855 6747471 94 + 10161210 --------CGUAAUCUAUAUGUUGCCCUCCACUGCAAACGGUUGUAGCUUUGGAUCAGGCUACAAUCGUAACGCCUAGGAGGGC-AUUUGAUCUAGCAACGCA---------------- --------(((...(((.((..((((((((...((..(((((((((((((......)))))))))))))...))...)))))))-)....)).)))..)))..---------------- ( -36.30, z-score = -4.15, R) >droMoj3.scaffold_6540 9778823 94 - 34148556 --------AUUGAUGCAGCUGUGGCCCUCCACUGCAAACGAUGGCACCUUGGAAUCAGGCUGCAAUCGUAACGCCUAGGAGGGC-AGUUUAUUUAGCAACGCA---------------- --------............((((((((((...((..(((((.(((.((((....)))).))).)))))...))...)))))))-.(((.....)))..))).---------------- ( -32.70, z-score = -1.35, R) >droGri2.scaffold_14906 8079987 96 + 14172833 ------UUUGU-AUAUAUAUAUAGCUCUCCACUGCAAACGGUUGGAGCUUUCGGUCAGGCUACAAACGUAACGCCUAGGAGAGCAAUUUGAUAUAGCAAUGCA---------------- ------.((((-.(((((..((.(((((((...((..(((.(((.(((((......))))).))).)))...))...))))))).))...)))))))))....---------------- ( -27.90, z-score = -2.26, R) >triCas2.ChLG5 2832875 96 + 18847211 -----------UUCGAAAGCAAUAUUUUUUUAUUUUCUCAAUUACGGCAAAACUGUACGAAACAAAGGAACUACUUUGAUGUGAACUUGUGUAAAAUGAUCCGGGGA------------ -----------..(....)...((((((..((..(((.((..(((((.....))))).....(((((......))))).)).)))..))...)))))).........------------ ( -11.80, z-score = 1.28, R) >consensus __________U_AUACAGCUGUUGCUCUUCAGUUACCACGAUGGAAGCUUUGAAUCAAGUUUCAAUCGUAACGCCUAGAAGAGC_AUUUUAUCUGGCAACGCAGGAA____________ .......................(((((((.......(((...(((((..........)))))...)))........)))))))................................... (-11.57 = -11.53 + -0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:30 2011