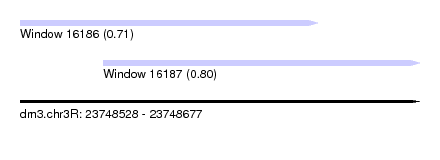
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,748,528 – 23,748,677 |
| Length | 149 |
| Max. P | 0.802668 |

| Location | 23,748,528 – 23,748,639 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.02 |
| Shannon entropy | 0.26223 |
| G+C content | 0.37864 |
| Mean single sequence MFE | -25.21 |
| Consensus MFE | -19.85 |
| Energy contribution | -19.48 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.707472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23748528 111 + 27905053 ------GCCAUUUCCGGCAGCUCUCUUUCUUUCUU---CGUUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAAUAUUUAGCACCUUUCAUUGGGUUUGAAAAUCAACGAAAA ------(((......)))...............((---((((((((((......))))))).(((.(((((....)))))...(((((.((((......))))))))).))).))))).. ( -25.70, z-score = -1.60, R) >droSim1.chr3R 23458529 111 + 27517382 ------GCCAUUUCCGGCAGCUCUCUUUCUUUCUU---CGUUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAAUAUAUAGCACCUUUCAUUGGGUUUGAAAAUCAACGAAAA ------(((......))).(((............(---(((.((((((......))))))))))..(((((....)))))......)))((((......))))................. ( -24.30, z-score = -1.25, R) >droSec1.super_22 373952 111 + 1066717 ------GCCAUUUCCGGCAGCUCUCUUUCUUUCUU---CGUUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAAUAUAUAGCACCUUUCAUUGGGUUUGAAAAUCAACGAAAA ------(((......))).(((............(---(((.((((((......))))))))))..(((((....)))))......)))((((......))))................. ( -24.30, z-score = -1.25, R) >droYak2.chr3R 6082878 111 - 28832112 ------GCCAUUUCCGGCAGCUCUCUUUCUUUCUU---CGUUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAAUAUUUAACACCUUUCAUUGGGUUUGAAAAUCAACGAAAA ------(((......)))...............((---((((((((((......))))))).(((.(((((....)))))...(((((.((((......))))))))).))).))))).. ( -25.60, z-score = -2.07, R) >droEre2.scaffold_4820 6176065 111 - 10470090 ------GCCAUUUCCGGCAGCUCUCUUUCUUUCUU---CGUUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAAUAUUUAACACCUUUCAUUGGGUUUGAAAAUCAACGAAAA ------(((......)))...............((---((((((((((......))))))).(((.(((((....)))))...(((((.((((......))))))))).))).))))).. ( -25.60, z-score = -2.07, R) >droAna3.scaffold_13340 21884612 117 + 23697760 UUUUCCGCCAUUUCCGGCAGCUCUCUUUCUUUCUU---CGUUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAAAAUUUAACACCAUUCAUUGGGCUUGAAAAUCAACGAAAA (((((.(((......)))(((((...........(---(((.((((((......))))))))))..(((((....)))))...................))))).))))).......... ( -26.10, z-score = -1.58, R) >dp4.chr2 22454123 114 - 30794189 ------GUCAUUUCCGGCGACCUUCCCUCUUUCUUUCUCGUUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAGUAUUUAAUUCCACUUAUUGGGCUUAAAAAUCAACGAACA ------(((......)))...................(((((((((((......))))))).(((.(((((....)))))...(((((..(((.....)))..))))).))).))))... ( -21.70, z-score = -0.62, R) >droPer1.super_0 9647827 114 - 11822988 ------GUCAUUUCCGGCGACCUUCCCUCUUUCUUUCUCGUUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAGUAUUUAAUUCCACUUAUUGGGCUUAAAAAUCAACGAACA ------(((......)))...................(((((((((((......))))))).(((.(((((....)))))...(((((..(((.....)))..))))).))).))))... ( -21.70, z-score = -0.62, R) >droWil1.scaffold_181130 1624497 115 - 16660200 UUUUCAGCCACUUCCGGCAGCUC--UUUCUUUCUU---CGUUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAAUAUAAAACACCAUUUAUUGGGCUUGAAAAUCAACAAAAA (((((((((......)))(((((--.........(---(((.((((((......))))))))))..(((((....)))))...................))))))))))).......... ( -27.80, z-score = -2.93, R) >droVir3.scaffold_12855 6746406 117 - 10161210 UUUUAGCCCAUUUCCGGCAGCUCUCUUUCUUUCUU---CGUUCUUCUUAUUAAAUAGAGGACGGCAUCUGCUUUUGCAGAAAAAUUAAAACCAUUUAUUGGGAUUAAAAAUCAGCGAAAA .....(((.......)))...........((((..---(((.(((((........))))))))((.(((((....)))))....((((..(((.....)))..))))......)))))). ( -26.00, z-score = -1.61, R) >droMoj3.scaffold_6540 9777777 117 + 34148556 UUUUAGGUCGUUUCCGGCAGCUCUCUUUCUUUCUU---CGUUCUUCUUAUUAAAUAGAGGACGGCAUCUGCGUUUGCAGAAAAAUUAAAACCAUUUAUUGGGAUUAAAAAUCAGCGAAAA ......((((....))))...........((((..---(((.(((((........))))))))((.(((((....)))))....((((..(((.....)))..))))......)))))). ( -25.00, z-score = -0.92, R) >droGri2.scaffold_14906 8078938 117 - 14172833 UUUUCGGCCAUUUCCGGCAGCUCUCUUUCUUUCUU---CGUUCUUCUUAUUAAAAAGAGGACGGCAUCUGCGUUUGCAGAAAAUUUAACACCAUUUAUUGGGAUUAAAAAUCAGCGAAAA .((((.(((......))).(((............(---(((.((((((......))))))))))..(((((....)))))...(((((..(((.....)))..)))))....))))))). ( -28.70, z-score = -2.15, R) >consensus ______GCCAUUUCCGGCAGCUCUCUUUCUUUCUU___CGUUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAAUAUUUAACACCAUUCAUUGGGUUUGAAAAUCAACGAAAA ......(((......)))...........((((.....(((.((((((......)))))))))((.(((((....))))).....(((..((.......))..))).......)))))). (-19.85 = -19.48 + -0.38)
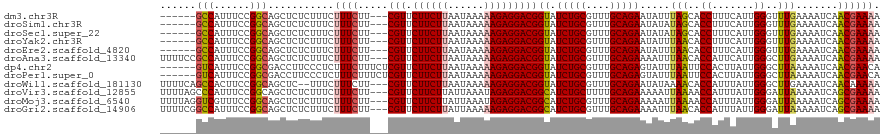
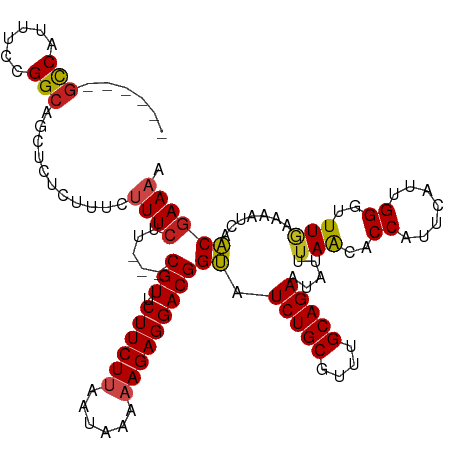
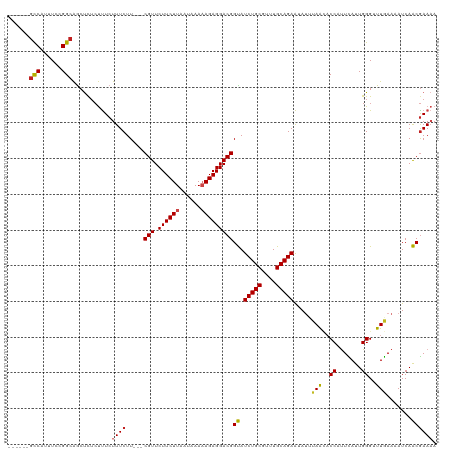
| Location | 23,748,559 – 23,748,677 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.92 |
| Shannon entropy | 0.41082 |
| G+C content | 0.40476 |
| Mean single sequence MFE | -29.11 |
| Consensus MFE | -18.95 |
| Energy contribution | -18.84 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.802668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23748559 118 + 27905053 UUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAAUAUUUAGCACCUUUCAUUGGGUUUGAAAAUCAACGAAAAUGGUGAGAAACGAAGCACAAGGCACUCCGCAACCGCAG-- .(((((((......)))))))......(((((.((((.((...(((..((((((((.((((.........)))).))))..))))..)))....((.....))..)).)))).)))))-- ( -28.90, z-score = -1.41, R) >droSim1.chr3R 23458560 118 + 27517382 UUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAAUAUAUAGCACCUUUCAUUGGGUUUGAAAAUCAACGAAAAUGGUGAGAAACGAAGCCCAAGGCACUCCGCAACCGCCG-- .(((((((......)))))))((((.(((((....))))).......((...((((((((..((((........))))..))))))))......(((...))).....)).))))...-- ( -29.40, z-score = -1.30, R) >droSec1.super_22 373983 118 + 1066717 UUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAAUAUAUAGCACCUUUCAUUGGGUUUGAAAAUCAACGAAAAUGGUGAGAAACGAAGCCCAAGGCACUCCGCAACCACCG-- .(((((((......)))))))((((.(((((....))))).......((...((((((((..((((........))))..))))))))......(((...))).....))....))))-- ( -29.00, z-score = -1.58, R) >droYak2.chr3R 6082909 118 - 28832112 UUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAAUAUUUAACACCUUUCAUUGGGUUUGAAAAUCAACGAAAAUGGUGAGAAACGACGCCCAAAGCACUCCGCAACCGCCU-- .(((((((......)))))))((((...((((..(((.(((............))).(((((((((....(((.(......).)))....))).)))))).)))...))))))))...-- ( -27.10, z-score = -1.13, R) >droEre2.scaffold_4820 6176096 118 - 10470090 UUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAAUAUUUAACACCUUUCAUUGGGUUUGAAAAUCAACGAAAAUGGUGAGAAACGACGCCCAAAGCACUCCGCAACCGCCG-- .(((((((......)))))))((((...((((..(((.(((............))).(((((((((....(((.(......).)))....))).)))))).)))...))))...))))-- ( -27.80, z-score = -1.30, R) >droAna3.scaffold_13340 21884649 120 + 23697760 UUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAAAAUUUAACACCAUUCAUUGGGCUUGAAAAUCAACGAAAAUGGUGAGAAACUAAGUGGACAAGGGCCCGCCACCGUCCCG ....((((......))))(((((((.(((((....)))))........((((((((.((((.........)))).))..)))))).........((((........)))).))))))).. ( -32.40, z-score = -1.73, R) >dp4.chr2 22454157 118 - 30794189 UUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAGUAUUUAAUUCCACUUAUUGGGCUUAAAAAUCAACGAACAUGGUGAGAAAGUGUGCAUCGUUAGUGCCUGGGCCGCCG-- .(((((((......)))))))((((.(((((....)))))....................((((((...((.(((((.((((.(.....).))))..))))).))...))))))))))-- ( -30.30, z-score = -0.87, R) >droPer1.super_0 9647861 118 - 11822988 UUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAGUAUUUAAUUCCACUUAUUGGGCUUAAAAAUCAACGAACAUGGUGAGAAAGUGUGCAUCGUUAGUGCCUGGGCCGCCG-- .(((((((......)))))))((((.(((((....)))))....................((((((...((.(((((.((((.(.....).))))..))))).))...))))))))))-- ( -30.30, z-score = -0.87, R) >droWil1.scaffold_181130 1624532 118 - 16660200 UUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAAUAUAAAACACCAUUUAUUGGGCUUGAAAAUCAACAAAAAUGGUAAGAAAAUGAACACUAACCACACCUGUUCCACCA-- .(((((((......))))))).(((.(((((....))))).........(((((((.(((....(....)....))))))))))........(((((...........))))).))).-- ( -24.10, z-score = -1.92, R) >droVir3.scaffold_12855 6746443 118 - 10161210 UUCUUCUUAUUAAAUAGAGGACGGCAUCUGCUUUUGCAGAAAAAUUAAAACCAUUUAUUGGGAUUAAAAAUCAGCGAAAAUGGUGAGAAAGUGUAUUUAAGUGUUGCUAGGGUCGCCG-- .((((((........))))))((((.(((((....)))))....((((..(((.....)))..))))........((...(((..(.(...((....))..).)..)))...))))))-- ( -28.70, z-score = -1.79, R) >droMoj3.scaffold_6540 9777814 118 + 34148556 UUCUUCUUAUUAAAUAGAGGACGGCAUCUGCGUUUGCAGAAAAAUUAAAACCAUUUAUUGGGAUUAAAAAUCAGCGAAAAUGGUGAGCAAAUGUGUUUGAGUGUCGUCCGUGCCACCG-- ....(((........)))((((((((((.((((((((.......((((..(((.....)))..))))...(((.(......).)))))))))))....)).)))))))).........-- ( -31.10, z-score = -2.03, R) >droGri2.scaffold_14906 8078975 118 - 14172833 UUCUUCUUAUUAAAAAGAGGACGGCAUCUGCGUUUGCAGAAAAUUUAACACCAUUUAUUGGGAUUAAAAAUCAGCGAAAAUGGUGAGAAAGUGUGCUCAAUGAAUGCCCAGAUCACAG-- .(((((((......))))))).(((((...((((.(((.(...(((..((((((((.(((.(((.....)))..)))))))))))..))).).)))..)))).)))))..........-- ( -30.20, z-score = -2.23, R) >consensus UUCUUCUUAAUAAAAAGAGGACGGUAUCUGCGUUUGCAGAAUAUUUAACACCAUUCAUUGGGUUUGAAAAUCAACGAAAAUGGUGAGAAACUAAGCACAAGGCACGCCGCAACCGCCG__ .(((((((......))))))).(((.(((((....)))))........((((.(((.((((.........)))).)))...))))..........................)))...... (-18.95 = -18.84 + -0.11)

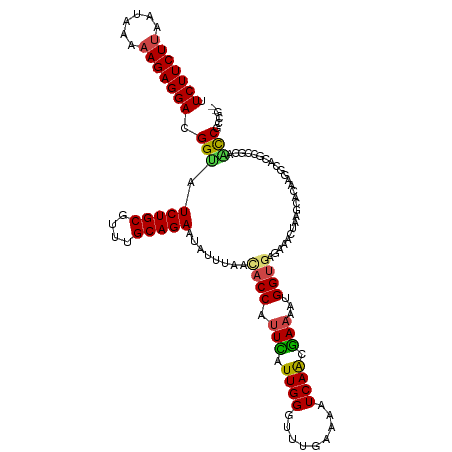
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:27 2011