| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,735,276 – 23,735,362 |
| Length | 86 |
| Max. P | 0.967835 |

| Location | 23,735,276 – 23,735,362 |
|---|---|
| Length | 86 |
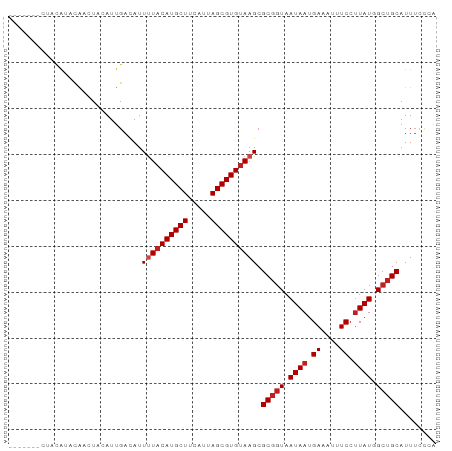
| Sequences | 12 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 88.72 |
| Shannon entropy | 0.24177 |
| G+C content | 0.37134 |
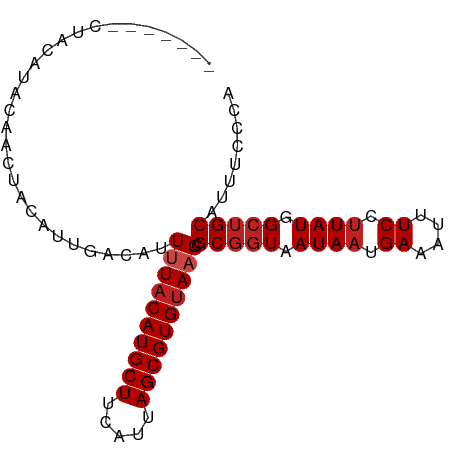
| Mean single sequence MFE | -19.58 |
| Consensus MFE | -18.17 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23735276 86 - 27905053 -------GUACAUACGACUACAUUGACAUUUUACAUGCUUCAUUAGCGUGCAAGCGCGGUAAUAAUGACAUUUCCUUAUGGCUGCAUUUCCCA -------..........................((((((.....)))))).....(((((.((((.((....)).)))).)))))........ ( -15.40, z-score = -0.10, R) >droSim1.chr3R 23443448 86 - 27517382 -------GUACAUACGACUACAUUGACAUUUUACAUGCUUCAUUAGCGUGUAAGCGCGGUAAUAAUGAAAUUUCCUUAUGGCUGCAUUUCCCA -------......................((((((((((.....)))))))))).(((((.((((.((....)).)))).)))))........ ( -19.80, z-score = -1.80, R) >droSec1.super_22 360595 86 - 1066717 -------GUACAUACGACUACAUUGACAUUUUACAUGCUUCAUUAGCGUGUAAGCGCGGUAAUAAUGAAAUUUCCUUAUGGCUGCAUUUCCCA -------......................((((((((((.....)))))))))).(((((.((((.((....)).)))).)))))........ ( -19.80, z-score = -1.80, R) >droEre2.scaffold_4820 6159975 86 + 10470090 -------AUAUGUACGACUACAUUGACAUUUUACAUGCUUCAUUAGCGUGUAAGCGCGGUAAUAAUGAAAUUUCCUUAUGGCUGCAUUUCUCA -------..(((((....)))))......((((((((((.....)))))))))).(((((.((((.((....)).)))).)))))........ ( -20.90, z-score = -2.03, R) >droYak2.chr3R 6068739 86 + 28832112 -------GUAUGUACGACUACAUUGACAUUUUACAUGCUUCAUUAGCGUGUAAGCGCGGUAAUAAUGAAAUUUCCUUAUGGCUGCAUUUCCCA -------(((((((....)))))..))..((((((((((.....)))))))))).(((((.((((.((....)).)))).)))))........ ( -21.20, z-score = -1.87, R) >droAna3.scaffold_13340 4730937 93 + 23697760 GUAUGUACGACAACUACAUACAUUGACAUUUUACAUGCUUCAUUAGCGUGUAAGCGCGGUAAUAAUGAAAUUUCCUUAUGGCUGCAUUUACAG (((((((.......)))))))........((((((((((.....)))))))))).(((((.((((.((....)).)))).)))))........ ( -26.00, z-score = -3.37, R) >dp4.chr2 4094342 87 - 30794189 -----CAC-ACAACUAUAUACAUUGAUAUUCUACAUGCUUCAUUAGCGUGUAAGCGCGGUAAUAAUGAAAUUUCCUUAUGGCUGCAUUUCCAG -----...-......................((((((((.....))))))))...(((((.((((.((....)).)))).)))))........ ( -18.50, z-score = -1.78, R) >droPer1.super_7 4184597 88 + 4445127 ----ACAC-ACAACUAUAUACAUUGAUAUUCUACAUGCUUCAUUAGCGUGUAAGCGCGGUAAUAAUGAAAUUUCCUUAUGGCUGCAUUUCCAG ----....-......................((((((((.....))))))))...(((((.((((.((....)).)))).)))))........ ( -18.50, z-score = -1.79, R) >droWil1.scaffold_181089 3987061 86 - 12369635 -------CUACAUACAACUACAUUGACAUUUUACAUGCUUCAUUAGCGUGUAAGCGCGGUAAUAAUGAAAUUUCCCUAUGGCUGCAUUUCCAA -------......................((((((((((.....)))))))))).(((((.(((..((....))..))).)))))........ ( -18.10, z-score = -1.72, R) >droMoj3.scaffold_6540 2055003 85 + 34148556 --------UACAUACAACUACAUUGACAUUUUACAUGCUUCAUUAGCGUGUAAGCGCGGUAAUAAUGAAAUUUCCUUAUGGCUGCAUUUGCCA --------.....................((((((((((.....)))))))))).(((((.((((.((....)).)))).)))))........ ( -19.80, z-score = -1.72, R) >droVir3.scaffold_12855 8346426 85 + 10161210 --------UACAUACAACUACAUUGACAUUUUACAUGCUUCAUUAGCGUGUAAGCGCGGUAAUAAUGAAAUUUCCUUAUGGCUGCAUUUUCAA --------..............((((...((((((((((.....)))))))))).(((((.((((.((....)).)))).)))))....)))) ( -20.60, z-score = -2.79, R) >droGri2.scaffold_14830 6064249 80 + 6267026 --------CACAUACAACUACAUUGACAUUUUACAUGCUUCAUUAGCGUGUAAGCGC--UAAUAAUGAAAUUUCCUUAUGGCUGCAUUGC--- --------......................(((((((((.....)))))))))((((--((.(((.((....)).))))))).)).....--- ( -16.40, z-score = -1.32, R) >consensus _______CUACAUACAACUACAUUGACAUUUUACAUGCUUCAUUAGCGUGUAAGCGCGGUAAUAAUGAAAUUUCCUUAUGGCUGCAUUUCCCA .............................((((((((((.....)))))))))).(((((.((((.((....)).)))).)))))........ (-18.17 = -18.68 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:25 2011