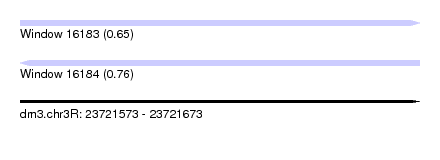
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,721,573 – 23,721,673 |
| Length | 100 |
| Max. P | 0.755994 |

| Location | 23,721,573 – 23,721,673 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.94 |
| Shannon entropy | 0.45561 |
| G+C content | 0.45359 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -18.32 |
| Energy contribution | -18.31 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.652062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
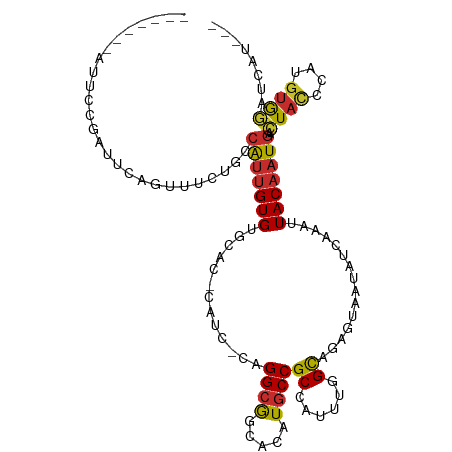
>dm3.chr3R 23721573 100 + 27905053 ---AUGAUUCACAUGGGUAGUCAUUGUAAUUUGAUAUUACUCUGCCAAAUGGCGGCAUGUGCCGCCUG-GAUG-GUGCACACAAUGGCAGAAACUGAAUCGGAAU------- ---.(((((((.....(((((((........))))..)))(((((((...((((((....))))))..-....-((....))..)))))))...)))))))....------- ( -33.20, z-score = -2.12, R) >droSim1.chr3R 23429394 100 + 27517382 ---AUGAUCCACAUGGAUAGUCAUUGUAAUUUGAUAUUACUCUGCCAAAUGGCGGCAUGUGCCGCCUG-GAUG-GUGCACACAAUGGCAGAAACUGAAUCGGAAU------- ---...((((....))))..(((..(((((.....)))))(((((((...((((((....))))))..-....-((....))..)))))))...)))........------- ( -30.60, z-score = -1.42, R) >droSec1.super_22 344962 100 + 1066717 ---AUGAUCCACAUGGAUAGUCAUUGUAAUUUGAUAUUACUCUGCCAAAUGGCGGCAUGUGCCGCCUG-GAUG-GUGCACACAAUGGCAGAAACUGAAUCGGAAU------- ---...((((....))))..(((..(((((.....)))))(((((((...((((((....))))))..-....-((....))..)))))))...)))........------- ( -30.60, z-score = -1.42, R) >droEre2.scaffold_4820 6145915 100 - 10470090 ---AUGAUCCACAUGGGUAGCUAUUGUAAUUUGAUAUUACUCUGCCAAAUGGCGGCAUGUGCCGCCUG-GAUG-GUGCACACAAUGGCAGAAACUGAAUCGAAAU------- ---...((((....))))...........((((((.....(((((((...((((((....))))))..-....-((....))..)))))))......))))))..------- ( -30.30, z-score = -1.48, R) >droYak2.chr3R 6054514 100 - 28832112 ---AUGAUCCACAUGGGUAGUCAUUGUAAUUUGAUAUUACUCUGCCAAAUGGCGGCAUGUGCCGCCUG-GAUG-GUGCACACAAUGGCAGAAACUGAAUCGGAAU------- ---....(((...(.(((.((((........)))).....(((((((...((((((....))))))..-....-((....))..))))))).))).)...)))..------- ( -30.60, z-score = -1.04, R) >droAna3.scaffold_13340 4716928 111 - 23697760 ACACUGAUCCACAUGGGUAGUCAUUGUAAGUUGAUAUUAUGCUGCGAAAUGGCGGCAUGUGUUGCCUG-GUGGUGCACACACAACGCCACAAUCGGAAUCAGAACCGAGCGA ...(((((((....(((((((((........))))..((((((((......))))))))...))))).-((((((.........))))))....)).))))).......... ( -34.60, z-score = -0.41, R) >dp4.chr2 4077173 80 + 30794189 ----AGAUCCACAUGGGUAGUCAUUGUAAUUUGAUAUUAUGCUGCGAAAUGGCGGCAUGUGUUGCCUC-------CGCACACAAUGGCAAU--------------------- ----..((((....)))).((((((((...........(((((((......)))))))((((......-------.))))))))))))...--------------------- ( -26.00, z-score = -1.43, R) >droPer1.super_7 4167745 80 - 4445127 ----AGAUCCACAUGGGUAGUCAUUGUAAUUUGAUAUUAUGCUGCGAAAUGGCGGCAUGUGUUGCCUC-------CGCACACAAUGGCAAU--------------------- ----..((((....)))).((((((((...........(((((((......)))))))((((......-------.))))))))))))...--------------------- ( -26.00, z-score = -1.43, R) >droWil1.scaffold_181089 3969408 82 + 12369635 --------AUGUAUGGGUAUUCAAAGUAAUUUGAUAUUAUGCUACGAAAUGGCGGCAUGUGUUCCCGAUGGUGGCUCCACACAAUGGCAU---------------------- --------.(((.((((...(((((....)))))......(((((.(..(((..((....))..))).).))))))))).))).......---------------------- ( -19.60, z-score = 0.39, R) >consensus ___AUGAUCCACAUGGGUAGUCAUUGUAAUUUGAUAUUACUCUGCCAAAUGGCGGCAUGUGCCGCCUG_GAUG_GUGCACACAAUGGCAGAAACUGAAUCGGAAU_______ ......((((....)))).((((((((.....(........)........((((((....))))))..............))))))))........................ (-18.32 = -18.31 + -0.01)



| Location | 23,721,573 – 23,721,673 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 75.94 |
| Shannon entropy | 0.45561 |
| G+C content | 0.45359 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -12.60 |
| Energy contribution | -12.52 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.755994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23721573 100 - 27905053 -------AUUCCGAUUCAGUUUCUGCCAUUGUGUGCAC-CAUC-CAGGCGGCACAUGCCGCCAUUUGGCAGAGUAAUAUCAAAUUACAAUGACUACCCAUGUGAAUCAU--- -------.....((((((...(((((((.((..((...-))..-))((((((....))))))...)))))))(((((.....)))))..............))))))..--- ( -34.10, z-score = -4.01, R) >droSim1.chr3R 23429394 100 - 27517382 -------AUUCCGAUUCAGUUUCUGCCAUUGUGUGCAC-CAUC-CAGGCGGCACAUGCCGCCAUUUGGCAGAGUAAUAUCAAAUUACAAUGACUAUCCAUGUGGAUCAU--- -------.....((((((...(((((((.((..((...-))..-))((((((....))))))...)))))))(((((.....)))))..............))))))..--- ( -33.60, z-score = -3.18, R) >droSec1.super_22 344962 100 - 1066717 -------AUUCCGAUUCAGUUUCUGCCAUUGUGUGCAC-CAUC-CAGGCGGCACAUGCCGCCAUUUGGCAGAGUAAUAUCAAAUUACAAUGACUAUCCAUGUGGAUCAU--- -------.....((((((...(((((((.((..((...-))..-))((((((....))))))...)))))))(((((.....)))))..............))))))..--- ( -33.60, z-score = -3.18, R) >droEre2.scaffold_4820 6145915 100 + 10470090 -------AUUUCGAUUCAGUUUCUGCCAUUGUGUGCAC-CAUC-CAGGCGGCACAUGCCGCCAUUUGGCAGAGUAAUAUCAAAUUACAAUAGCUACCCAUGUGGAUCAU--- -------.....((((((...(((((((.((..((...-))..-))((((((....))))))...)))))))(((((.....)))))..............))))))..--- ( -33.60, z-score = -3.49, R) >droYak2.chr3R 6054514 100 + 28832112 -------AUUCCGAUUCAGUUUCUGCCAUUGUGUGCAC-CAUC-CAGGCGGCACAUGCCGCCAUUUGGCAGAGUAAUAUCAAAUUACAAUGACUACCCAUGUGGAUCAU--- -------.....((((((...(((((((.((..((...-))..-))((((((....))))))...)))))))(((((.....)))))..............))))))..--- ( -33.60, z-score = -3.28, R) >droAna3.scaffold_13340 4716928 111 + 23697760 UCGCUCGGUUCUGAUUCCGAUUGUGGCGUUGUGUGUGCACCAC-CAGGCAACACAUGCCGCCAUUUCGCAGCAUAAUAUCAACUUACAAUGACUACCCAUGUGGAUCAGUGU ..((((((........))((..((((((..((((((((.....-...)).))))))..)))))).))).))).............(((.(((((((....)))).))).))) ( -31.80, z-score = -0.44, R) >dp4.chr2 4077173 80 - 30794189 ---------------------AUUGCCAUUGUGUGCG-------GAGGCAACACAUGCCGCCAUUUCGCAGCAUAAUAUCAAAUUACAAUGACUACCCAUGUGGAUCU---- ---------------------.(((..((((((((((-------((((((.....)))).....)))))).))))))..)))........((((((....)))).)).---- ( -18.80, z-score = -0.11, R) >droPer1.super_7 4167745 80 + 4445127 ---------------------AUUGCCAUUGUGUGCG-------GAGGCAACACAUGCCGCCAUUUCGCAGCAUAAUAUCAAAUUACAAUGACUACCCAUGUGGAUCU---- ---------------------.(((..((((((((((-------((((((.....)))).....)))))).))))))..)))........((((((....)))).)).---- ( -18.80, z-score = -0.11, R) >droWil1.scaffold_181089 3969408 82 - 12369635 ----------------------AUGCCAUUGUGUGGAGCCACCAUCGGGAACACAUGCCGCCAUUUCGUAGCAUAAUAUCAAAUUACUUUGAAUACCCAUACAU-------- ----------------------.......(((((((..((......))......((((..(......)..))))....(((((....)))))....))))))).-------- ( -15.80, z-score = -0.19, R) >consensus _______AUUCCGAUUCAGUUUCUGCCAUUGUGUGCAC_CAUC_CAGGCGGCACAUGCCGCCAUUUGGCAGAGUAAUAUCAAAUUACAAUGACUACCCAUGUGGAUCAU___ ..........................(((((((.............((((.....))))((......))...............))))))).((((....))))........ (-12.60 = -12.52 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:24 2011