| Sequence ID | dm3.chr3R |
|---|---|
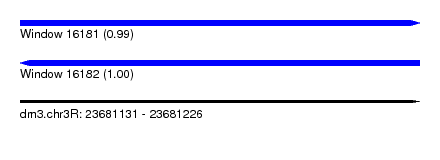
| Location | 23,681,131 – 23,681,226 |
| Length | 95 |
| Max. P | 0.997744 |

| Location | 23,681,131 – 23,681,226 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 87.41 |
| Shannon entropy | 0.20178 |
| G+C content | 0.44134 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -24.49 |
| Energy contribution | -26.55 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.988991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
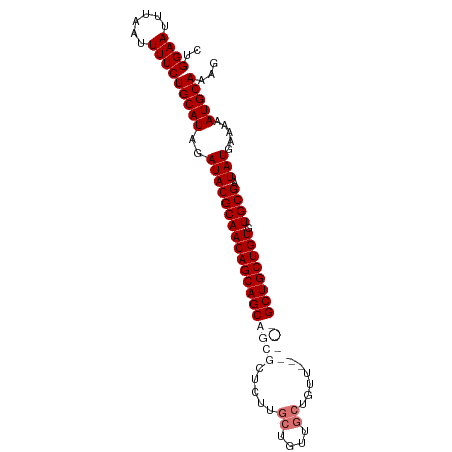
>dm3.chr3R 23681131 95 + 27905053 CUGGAAUUUAAUUUUCUGCAUAGAUACGCAACAGCAGCAGCGCUCUUGCUGUUGCUGUUCCUGCUGCUGCUGUGUGCGAUAUGAAAAAUGCAAAG ..((((......))))(((((..(((((((((((((((((((.....((....))......)))))))))))).)))).))).....)))))... ( -33.50, z-score = -2.71, R) >droSim1.chr3R 23402985 95 + 27517382 CUGGAAUUUAAUUUUCUGCAUAGAUACGCAACAGCAGCAGCGCUCUUGCUGUUGCUGUUGCUGUUGCUGCUGUGUGCGAUAUGAAAAAUGCAAAG ..((((......))))(((((......(((((((((((((((....)))))))))))))))((((((........))))))......)))))... ( -37.20, z-score = -3.70, R) >droSec1.super_22 318525 89 + 1066717 CUGGAAUUUAAUUUUCUGCAUAGAUACGCAACAGCAGCAGCGCUCUUGCUGUUGCUGUU------GCUGCUGUGUGCGAUAUGAAAAAUGCAAAG ..((((......))))(((((......(((((((((((((((....)))))))))))))------))(((.....))).........)))))... ( -33.70, z-score = -3.37, R) >droYak2.chr3R 6026685 74 - 28832112 CUGGAAUUUAAUUUUCUGCAUAGAUACGCAACAGCAGCAGCGCU---------------------GCUGCUGUGUGCGAUAUGAAAAAUGCAAAG ..((((......))))(((((..((((((((((((((((....)---------------------)))))))).)))).))).....)))))... ( -25.50, z-score = -2.72, R) >consensus CUGGAAUUUAAUUUUCUGCAUAGAUACGCAACAGCAGCAGCGCUCUUGCUGUUGCUGUU______GCUGCUGUGUGCGAUAUGAAAAAUGCAAAG ..((((......))))(((((..(((((((((((((((((((.....((....))......)))))))))))).)))).))).....)))))... (-24.49 = -26.55 + 2.06)

| Location | 23,681,131 – 23,681,226 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 87.41 |
| Shannon entropy | 0.20178 |
| G+C content | 0.44134 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -23.83 |
| Energy contribution | -23.96 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.997744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
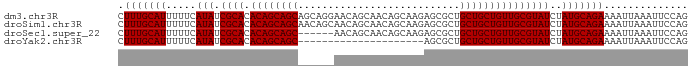
>dm3.chr3R 23681131 95 - 27905053 CUUUGCAUUUUUCAUAUCGCACACAGCAGCAGCAGGAACAGCAACAGCAAGAGCGCUGCUGCUGUUGCGUAUCUAUGCAGAAAAUUAAAUUCCAG .(((((((.....(((.((((.(((((((((((.(......)....((....))))))))))))))))))))..))))))).............. ( -33.90, z-score = -3.71, R) >droSim1.chr3R 23402985 95 - 27517382 CUUUGCAUUUUUCAUAUCGCACACAGCAGCAACAGCAACAGCAACAGCAAGAGCGCUGCUGCUGUUGCGUAUCUAUGCAGAAAAUUAAAUUCCAG .(((((((.....(((.((((.(((((((((.(.((....))....((....))).))))))))))))))))..))))))).............. ( -30.60, z-score = -3.17, R) >droSec1.super_22 318525 89 - 1066717 CUUUGCAUUUUUCAUAUCGCACACAGCAGC------AACAGCAACAGCAAGAGCGCUGCUGCUGUUGCGUAUCUAUGCAGAAAAUUAAAUUCCAG .(((((((....(.....((.....)).((------(((((((.((((......)))).)))))))))).....))))))).............. ( -30.30, z-score = -3.61, R) >droYak2.chr3R 6026685 74 + 28832112 CUUUGCAUUUUUCAUAUCGCACACAGCAGC---------------------AGCGCUGCUGCUGUUGCGUAUCUAUGCAGAAAAUUAAAUUCCAG .(((((((.....(((.((((.((((((((---------------------(....))))))))))))))))..))))))).............. ( -27.50, z-score = -4.52, R) >consensus CUUUGCAUUUUUCAUAUCGCACACAGCAGC______AACAGCAACAGCAAGAGCGCUGCUGCUGUUGCGUAUCUAUGCAGAAAAUUAAAUUCCAG .(((((((.....(((.((((.((((((((..........((....)).........)))))))))))))))..))))))).............. (-23.83 = -23.96 + 0.13)
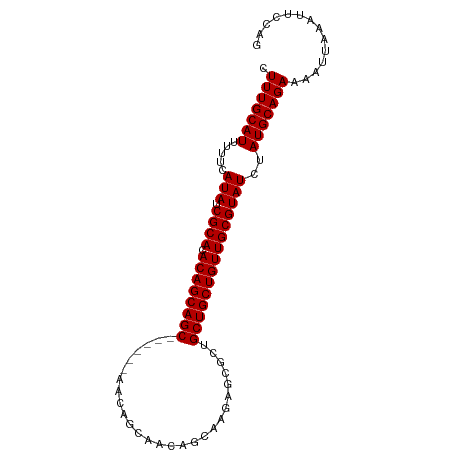
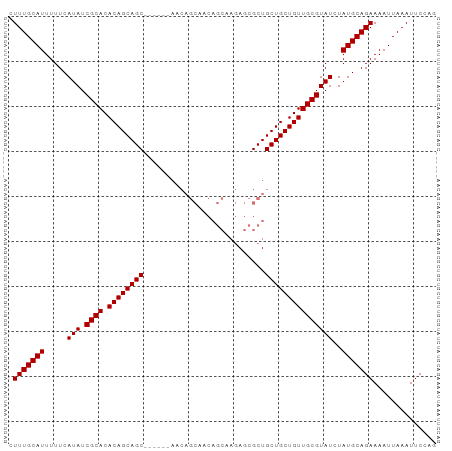
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:23 2011