| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,664,375 – 23,664,480 |
| Length | 105 |
| Max. P | 0.813114 |

| Location | 23,664,375 – 23,664,480 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 67.48 |
| Shannon entropy | 0.62184 |
| G+C content | 0.58178 |
| Mean single sequence MFE | -41.55 |
| Consensus MFE | -13.02 |
| Energy contribution | -13.05 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.813114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23664375 105 + 27905053 -----GCUGCUGCAGCUGUAUUCGAUUUCAAGGACACUCGAUGGGGCAGCUGCUGUAUCUCCAUGCUCAU------CCCCAUUCUCAUCGGUGUCCGGAUGCCGGCUGCUGGUAGC -----(((((((((((((((((((....)..((((((.(((((((((....)).((((....))))....------.......)))))))))))))))))).))))))).)))))) ( -45.80, z-score = -3.19, R) >droAna3.scaffold_13340 4670158 84 - 23697760 --------CCAGCUGCUGUAUUUGAUUUCAAGGACACUCGAUGGGGCAGCUGGCC-AUGGUCCUGCUCAU------CCUGGCUGCUACUAG---CUGGUAGC-------------- --------.((((((((.((((.((............)))))).))))))))(((-(.((..........------)))))).((((((..---..))))))-------------- ( -28.90, z-score = -0.14, R) >droEre2.scaffold_4820 6093691 102 - 10470090 --------GCUGCAGCUGUAUUCGAUUUCAAGGACACUCGAUGGGGCAGCUGCCCCAUCCUCGUGCUCAU------CCCCAUCCUCAUCGGUGUCCGGAUGCCGGCUGCUGGUAGC --------((((((((((((((((....)..(((((((.((((((((....))))))))...(((.....------...))).......)))))))))))).))))))).)))... ( -42.60, z-score = -2.34, R) >droYak2.chr3R 6008406 111 - 28832112 -----GCUGCUGCAGCUGUAUUCGAUUUCAAGGACACUCGAUGGGGCAGCUGCCCUAUCCCCAUGCUCAUGCUCAUCCGCAUCCUCAUCGGUGUCCGGAUGCCGACUGCUGGUAGC -----(((((((((((.(((((((....)..(((((((.((((((((....))))))))...(((...((((......))))...))).))))))))))))).).)))).)))))) ( -47.20, z-score = -3.02, R) >droSec1.super_22 302060 105 + 1066717 -----GCUGCUGCAGCUGUAUUCGAUUUCAAGGACACUCGAUGGGGCAGCUGCUCUAUCCCCAUGCUCAU------CCCCAUCCUCAUCGGUGUCCGGAUGCCGGCUGCUUGUAGC -----(((((.(((((((((((((....)..(((((((.(((((((.(((..............)))...------)))))))......)))))))))))).)))))))..))))) ( -45.34, z-score = -3.57, R) >droSim1.chr3R 23385612 105 + 27517382 -----GCUGCUGCAGCUGUAUUCGAUUUCAAGGACACUCGAUGGGGCAGCUGCUCUAUCCCCAUGCUCAU------CCCCAUCCUCAUCGGUGUCCGGAUGCCGGCUGCUGGUAGC -----(((((((((((((((((((....)..(((((((.(((((((.(((..............)))...------)))))))......)))))))))))).))))))).)))))) ( -46.24, z-score = -3.30, R) >dp4.chr2 4022945 100 + 30794189 CACUUGCAGCAGCAGCUGUAUUCGAUUUCAAGGACACUCGAUGGGGCAGCUGCCUCAUCCGAUGCUACUG-------CUGGU-GGUGCUGCUGGUGGUGGUG--GUGGCC------ ....((((((....))))))...........((.((((.((((((((....))))))))....(((((..-------(..((-......))..)..))))))--))).))------ ( -39.80, z-score = -0.56, R) >droGri2.scaffold_14830 5986251 95 - 6267026 -----GCAUUUGGAGCACUAGUCAGUUUCGAGAACACUCAAUGGGGCAGCUGCCUCAUC--ACAGCAGAG-------CUGGCCGAUGCUGGCGAUGCUGCUGCUGUAGC------- -----((.((((((((........))))))))........(((((((....))))))).--(((((((((-------((.((((....)))).).))).))))))).))------- ( -36.50, z-score = -0.55, R) >consensus _____GCUGCUGCAGCUGUAUUCGAUUUCAAGGACACUCGAUGGGGCAGCUGCCCUAUCCCCAUGCUCAU______CCCCAUCCUCAUCGGUGUCCGGAUGCCGGCUGCUGGUAGC ...........(((.(((...(((((....((....)).((((((((....))))))))...........................)))))....))).))).............. (-13.02 = -13.05 + 0.03)

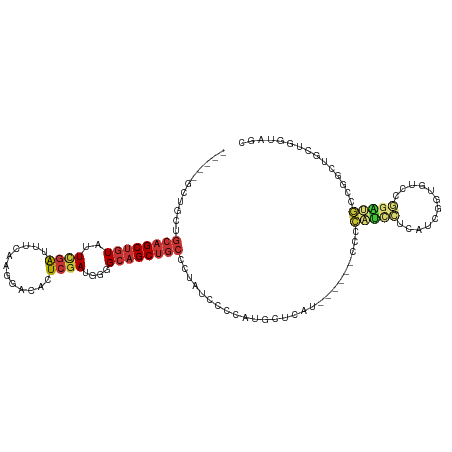

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:21 2011