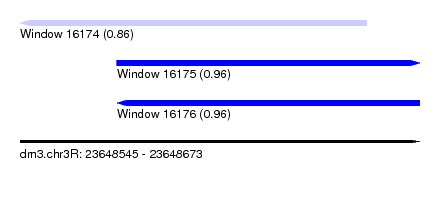
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,648,545 – 23,648,673 |
| Length | 128 |
| Max. P | 0.956516 |

| Location | 23,648,545 – 23,648,656 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.81 |
| Shannon entropy | 0.13176 |
| G+C content | 0.31233 |
| Mean single sequence MFE | -23.75 |
| Consensus MFE | -20.77 |
| Energy contribution | -20.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23648545 111 - 27905053 CAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGCGUGAGCAU---------GUCGGCAUAAAAACCAAUGGCAG .(((((((((((((.......(((((((....)).)))))(((......)))....))))))))))))).......((((....))))---------...(.(((........))).).. ( -24.80, z-score = -2.38, R) >droPer1.super_7 4098419 111 + 4445127 CAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUGUAAUUUAAUUUCUUAAUUAAAAUGCGAGAGCAU---------GUCUGCAUAAAAACCAAUGGAGG .(((((((((((((.......(((((((....)).)))))(((((.....).))))))))))))))))).......((((....))))---------..((.(((........))).)). ( -24.40, z-score = -2.25, R) >dp4.chr2 4005801 111 - 30794189 CAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUGUAAUUUAAUUUCUUAAUUAAAAUGCGAGAGCAU---------GUCUGCAUAAAAACCAAUGGAGG .(((((((((((((.......(((((((....)).)))))(((((.....).))))))))))))))))).......((((....))))---------..((.(((........))).)). ( -24.40, z-score = -2.25, R) >droAna3.scaffold_13340 4654057 111 + 23697760 CAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGCGCGAGCAU---------GUCGGCAUAAAAACCAAUGGCUG .(((((((((((((.......(((((((....)).)))))(((......)))....))))))))))))).......((((....))))---------...(.(((........))).).. ( -23.80, z-score = -1.72, R) >droEre2.scaffold_4820 6077983 111 + 10470090 CAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGCGUGAGCAU---------GUCGGCAUAAAAACCAAUGGCAG .(((((((((((((.......(((((((....)).)))))(((......)))....))))))))))))).......((((....))))---------...(.(((........))).).. ( -24.80, z-score = -2.38, R) >droYak2.chr3R 5991979 111 + 28832112 CAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGCGUAAGCAU---------GUCGGCAUAAAAACCAAUGGCAG .(((((((((((((.......(((((((....)).)))))(((......)))....))))))))))))).......((((....))))---------...(.(((........))).).. ( -23.80, z-score = -2.35, R) >droSec1.super_22 286182 111 - 1066717 CAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGCGUGAGCAU---------GUCGGCAUAAAAACCAAUGGCAG .(((((((((((((.......(((((((....)).)))))(((......)))....))))))))))))).......((((....))))---------...(.(((........))).).. ( -24.80, z-score = -2.38, R) >droSim1.chr3R 23369386 111 - 27517382 CAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGCGUGAGCAU---------GUCGGCAUAAAAACCAAUGGCAG .(((((((((((((.......(((((((....)).)))))(((......)))....))))))))))))).......((((....))))---------...(.(((........))).).. ( -24.80, z-score = -2.38, R) >droGri2.scaffold_14830 5966405 111 + 6267026 CAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGCGCGGGCAU---------GUCGGCAUAAAAUCCAACGACCG .(((((((((((((.......(((((((....)).)))))(((......)))....))))))))))))).......((((....))))---------((((.............)))).. ( -22.42, z-score = -1.30, R) >droVir3.scaffold_12855 8239201 109 + 10161210 CAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGCGCGGGCAU---------GUCGGCAUAAAACCCAAUGAC-- .(((((((((((((.......(((((((....)).)))))(((......)))....))))))))))))).......((((.(((....---------.))))))).............-- ( -21.10, z-score = -1.06, R) >droMoj3.scaffold_6540 1940168 109 + 34148556 CAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGCGCGGGCAU---------GUCGGCAUAAAAUCCAACGAC-- .(((((((((((((.......(((((((....)).)))))(((......)))....))))))))))))).......((((....))))---------((((.............))))-- ( -21.62, z-score = -1.35, R) >droWil1.scaffold_181089 3904546 118 - 12369635 CAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGCGUGAGCAUGCGCCAAUCGCCUGUAUGAAAAUCAACGAC-- .(((((((((((((.......(((((((....)).)))))(((......)))....))))))))))))).......((((....)))).......(((..((.((....)))).))).-- ( -24.30, z-score = -1.40, R) >consensus CAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGCGUGAGCAU_________GUCGGCAUAAAAACCAAUGGCAG .(((((((((((((.......(((((((....)).)))))(((......)))....))))))))))))).......((((....))))................................ (-20.77 = -20.77 + 0.00)

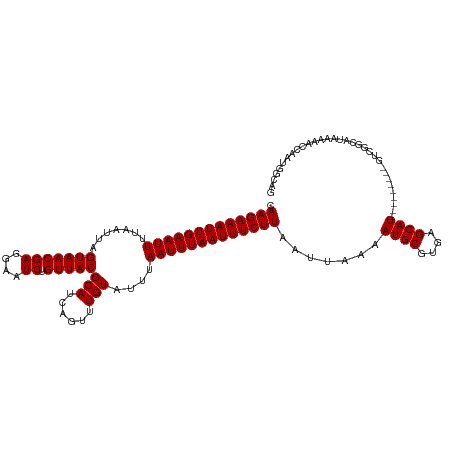
| Location | 23,648,576 – 23,648,673 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 88.94 |
| Shannon entropy | 0.23584 |
| G+C content | 0.28898 |
| Mean single sequence MFE | -18.24 |
| Consensus MFE | -16.29 |
| Energy contribution | -16.18 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23648576 97 + 27905053 GCAUUUUAAUUAAGAAAUUAAAUUAAAUACAAACUGAUGCGUAACAGAUUCCUCGUUACUAAUUAAAAUUCAAUUUCUUGCCGUUAAAAUGAGCAGC-------------------- .((((((((((((((((((.((((.....((......)).(((((((....)).))))).......)))).)))))))))..)))))))))......-------------------- ( -16.60, z-score = -1.89, R) >droPer1.super_7 4098450 115 - 4445127 GCAUUUUAAUUAAGAAAUUAAAUUACAUACAAACUGAUGCGUAACAGAUUCCUCGUUACUAAUUAAAAUUCAAUUUCUUGUCAUUAAAAUGAGCAGCAAAAGCAUCAGCGGCGGC-- .((((((((((((((((((.((((.(((........))).(((((((....)).))))).......)))).)))))))))..))))))))).((.((....((....)).)).))-- ( -22.30, z-score = -1.92, R) >dp4.chr2 4005832 115 + 30794189 GCAUUUUAAUUAAGAAAUUAAAUUACAUACAAACUGAUGCGUAACAGAUUCCUCGUUACUAAUUAAAAUUCAAUUUCUUGUCAUUAAAAUGAGCAGCAAAAGCAUCAGUGGCGGC-- ((.......((((....))))...........(((((((((((((((....)).)))))..............(((.(((((((....)))).))).))).)))))))).))...-- ( -23.10, z-score = -2.19, R) >droAna3.scaffold_13340 4654088 105 - 23697760 GCAUUUUAAUUAAGAAAUUAAAUUAAAUACAAACUGAUGCGUAACAGAUUCCUCGUUACUAAUUAAAAUUCAAUUUCUUGCCAUUAAAAUGAACAGCAGGGCUGU------------ .((((((((((((((((((.((((.....((......)).(((((((....)).))))).......)))).)))))))))..))))))))).(((((...)))))------------ ( -20.20, z-score = -2.35, R) >droEre2.scaffold_4820 6078014 97 - 10470090 GCAUUUUAAUUAAGAAAUUAAAUUAAAUACAAACUGAUGCGUAACAGAUUCCUCGUUACUAAUUAAAAUUCAAUUUCUUGCCAUUAAAAUGAGCAGC-------------------- .((((((((((((((((((.((((.....((......)).(((((((....)).))))).......)))).)))))))))..)))))))))......-------------------- ( -16.30, z-score = -2.05, R) >droYak2.chr3R 5992010 100 - 28832112 GCAUUUUAAUUAAGAAAUUAAAUUAAAUACAAACUGAUGCGUAACAGAUUCCUCGUUACUAAUUAAAAUUCAAUUUCUUGCCAUUAAAAUGAGCAGCCGC----------------- .((((((((((((((((((.((((.....((......)).(((((((....)).))))).......)))).)))))))))..))))))))).........----------------- ( -16.30, z-score = -1.57, R) >droSec1.super_22 286213 97 + 1066717 GCAUUUUAAUUAAGAAAUUAAAUUAAAUACAAACUGAUGCGUAACAGAUUCCUCGUUACUAAUUAAAAUUCAAUUUCUUGCCGUUAAAAUGAGCAGC-------------------- .((((((((((((((((((.((((.....((......)).(((((((....)).))))).......)))).)))))))))..)))))))))......-------------------- ( -16.60, z-score = -1.89, R) >droSim1.chr3R 23369417 96 + 27517382 GCAUUUUAAUUAAGAAAUUAAAUUAAAUACAAACUGAUGCGUAACAGAUUCCUCGUUACUAAUUAAAAUUCAAUUUCUUGCCGU-AAAAUGAGCAGC-------------------- .(((((((..(((((((((.((((.....((......)).(((((((....)).))))).......)))).)))))))))...)-))))))......-------------------- ( -15.50, z-score = -1.50, R) >droVir3.scaffold_12855 8239230 103 - 10161210 GCAUUUUAAUUAAGAAAUUAAAUUAAAUACAAACUGAUGCGUAACAGAUUCCUCGUUACUAAUUAAAAUUCAAUUUCUUGCCAUUAAAAUGAGCAGCGACAAC-------------- .((((((((((((((((((.((((.....((......)).(((((((....)).))))).......)))).)))))))))..)))))))))............-------------- ( -16.30, z-score = -1.91, R) >droMoj3.scaffold_6540 1940197 103 - 34148556 GCAUUUUAAUUAAGAAAUUAAAUUAAAUACAAACUGAUGCGUAACAGAUUCCUCGUUACUAAUUAAAAUUCAAUUUCUUGCCAUUAAAAUGAGCAGCACCAGA-------------- ....(((((((((....)).)))))))......(((.((((((((((....)).)))))...................((((((....))).)))))).))).-------------- ( -17.50, z-score = -2.34, R) >droWil1.scaffold_181089 3904584 117 + 12369635 GCAUUUUAAUUAAGAAAUUAAAUUAAAUACAAACUGAUGCGUAACAGAUUCCUCGUUACUAAUUAAAAUUCAAUUUCUUGCCAUUAAAAUGAGCAGCAGCAGUUCGUUCCUUGUUCA ....(((((((((....)).))))))).((.(((((.((((((((((....)).)))))...................((((((....))).)))))).))))).)).......... ( -19.90, z-score = -1.51, R) >consensus GCAUUUUAAUUAAGAAAUUAAAUUAAAUACAAACUGAUGCGUAACAGAUUCCUCGUUACUAAUUAAAAUUCAAUUUCUUGCCAUUAAAAUGAGCAGCA___________________ .((((((((((((((((((.((((.....((......)).(((((((....)).))))).......)))).)))))))))..))))))))).......................... (-16.29 = -16.18 + -0.11)

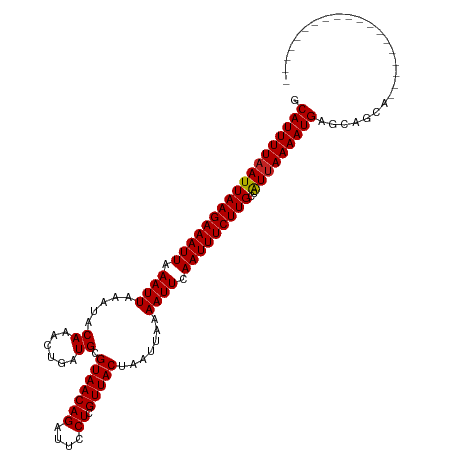
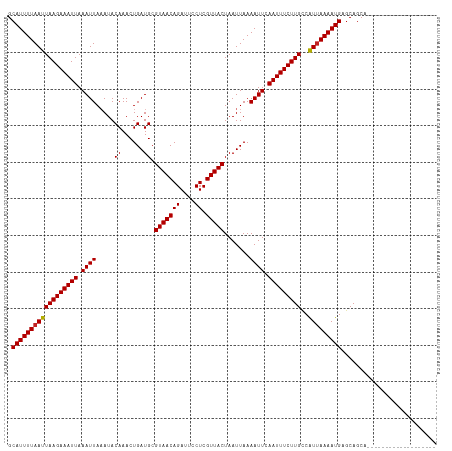
| Location | 23,648,576 – 23,648,673 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 88.94 |
| Shannon entropy | 0.23584 |
| G+C content | 0.28898 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -19.31 |
| Energy contribution | -19.40 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23648576 97 - 27905053 --------------------GCUGCUCAUUUUAACGGCAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGC --------------------......((((((((....(((((((((((((.......(((((((....)).)))))(((......)))....)))))))))))))..)))))))). ( -19.60, z-score = -1.83, R) >droPer1.super_7 4098450 115 + 4445127 --GCCGCCGCUGAUGCUUUUGCUGCUCAUUUUAAUGACAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUGUAAUUUAAUUUCUUAAUUAAAAUGC --((.((.((....))....)).)).(((((((((...(((((((((((((.......(((((((....)).)))))(((((.....).))))))))))))))))).))))))))). ( -26.40, z-score = -2.09, R) >dp4.chr2 4005832 115 - 30794189 --GCCGCCACUGAUGCUUUUGCUGCUCAUUUUAAUGACAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUGUAAUUUAAUUUCUUAAUUAAAAUGC --((.((.............)).)).(((((((((...(((((((((((((.......(((((((....)).)))))(((((.....).))))))))))))))))).))))))))). ( -23.92, z-score = -1.59, R) >droAna3.scaffold_13340 4654088 105 + 23697760 ------------ACAGCCCUGCUGUUCAUUUUAAUGGCAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGC ------------(((((...))))).(((((((((...(((((((((((((.......(((((((....)).)))))(((......)))....))))))))))))).))))))))). ( -25.80, z-score = -3.30, R) >droEre2.scaffold_4820 6078014 97 + 10470090 --------------------GCUGCUCAUUUUAAUGGCAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGC --------------------......(((((((((...(((((((((((((.......(((((((....)).)))))(((......)))....))))))))))))).))))))))). ( -22.10, z-score = -2.67, R) >droYak2.chr3R 5992010 100 + 28832112 -----------------GCGGCUGCUCAUUUUAAUGGCAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGC -----------------((....)).(((((((((...(((((((((((((.......(((((((....)).)))))(((......)))....))))))))))))).))))))))). ( -22.20, z-score = -1.99, R) >droSec1.super_22 286213 97 - 1066717 --------------------GCUGCUCAUUUUAACGGCAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGC --------------------......((((((((....(((((((((((((.......(((((((....)).)))))(((......)))....)))))))))))))..)))))))). ( -19.60, z-score = -1.83, R) >droSim1.chr3R 23369417 96 - 27517382 --------------------GCUGCUCAUUUU-ACGGCAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGC --------------------((((........-.))))(((((((((((((.......(((((((....)).)))))(((......)))....)))))))))))))........... ( -19.20, z-score = -1.70, R) >droVir3.scaffold_12855 8239230 103 + 10161210 --------------GUUGUCGCUGCUCAUUUUAAUGGCAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGC --------------............(((((((((...(((((((((((((.......(((((((....)).)))))(((......)))....))))))))))))).))))))))). ( -22.10, z-score = -2.04, R) >droMoj3.scaffold_6540 1940197 103 + 34148556 --------------UCUGGUGCUGCUCAUUUUAAUGGCAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGC --------------............(((((((((...(((((((((((((.......(((((((....)).)))))(((......)))....))))))))))))).))))))))). ( -22.10, z-score = -2.02, R) >droWil1.scaffold_181089 3904584 117 - 12369635 UGAACAAGGAACGAACUGCUGCUGCUCAUUUUAAUGGCAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGC ..........((((((((.((((((.(((....))))))...................(((((((....)).)))))))).))))))))......(((((((....))))))).... ( -24.10, z-score = -1.47, R) >consensus ___________________UGCUGCUCAUUUUAAUGGCAAGAAAUUGAAUUUUAAUUAGUAACGAGGAAUCUGUUACGCAUCAGUUUGUAUUUAAUUUAAUUUCUUAAUUAAAAUGC ..........................((((((((....(((((((((((((.......(((((((....)).)))))(((......)))....)))))))))))))..)))))))). (-19.31 = -19.40 + 0.09)

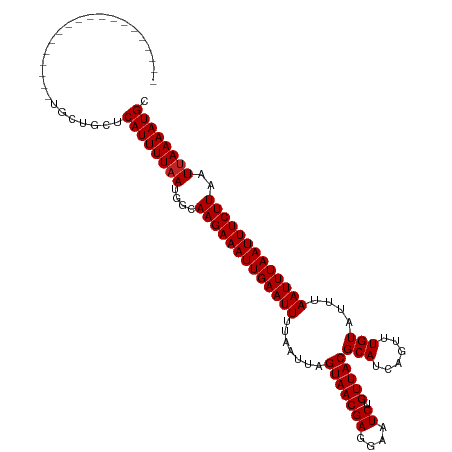
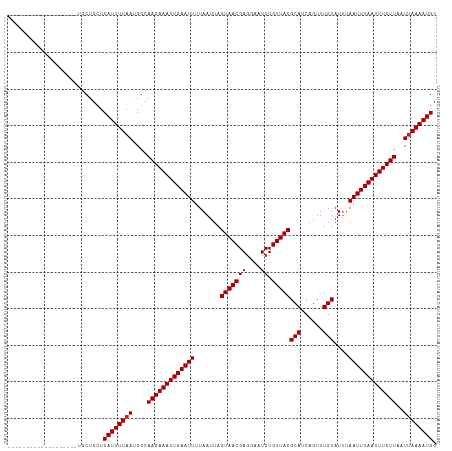
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:18 2011