
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,630,613 – 23,630,705 |
| Length | 92 |
| Max. P | 0.576265 |

| Location | 23,630,613 – 23,630,705 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 63.53 |
| Shannon entropy | 0.74531 |
| G+C content | 0.37394 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -4.88 |
| Energy contribution | -5.04 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.576265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
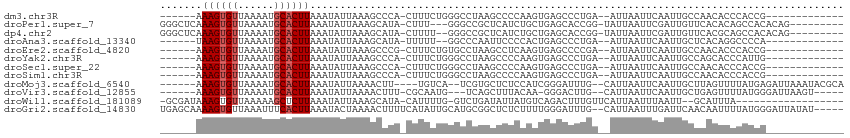
>dm3.chr3R 23630613 92 + 27905053 ------AAAGUGUUAAAAUGCACUUAAAUAUUAAAGCCCA-CUUUCUGGGCCUAAGCCCCAAGUGAGCCCUGA--AUUAAUUCAAUUGCCAACACCCACCG------------- ------.((((((......))))))..........((.((-(((...((((....)))).))))).))..(((--(....)))).................------------- ( -19.60, z-score = -2.71, R) >droPer1.super_7 4078923 100 - 4445127 GGGCUCAAAGUGUUAAAAUGCACUUAAAUAUUAAAGCAUA-CUUU---GGGCCGCUCAUCUGCUGAGCACCGG-UAUUAAUUCGAUUGUUCACACAGCCACACAG--------- .((((((((((......((((..............)))))-))))---)))))(((....((.((((((.(((-.......)))..)))))))).))).......--------- ( -24.24, z-score = -1.45, R) >dp4.chr2 3986202 101 + 30794189 GGGCUCAAAGUGUUAAAAUGCACUUAAAUAUUAAAGCAUA-CUUUU--GGGCCGCUCAUCUGCUGAGCACCGG-UAUUAAUUCGAUUGUUCACGCAGCCACACAG--------- .((((((((((((......)))))).......((((....-)))))--)))))......((((((((((.(((-.......)))..)))))).))))........--------- ( -27.10, z-score = -1.95, R) >droAna3.scaffold_13340 4633827 90 - 23697760 ------UAAGUGUUAAAAUGCACUUAAAUAUUAAAGCAUA-UUUUU--GGCCCAAUUCCCCACUGAGCCCUGA--AUUAAUUCAAUUGCUCACAGGCCCCA------------- ------(((((((......)))))))..............-.....--((((...........(((((..(((--(....))))...)))))..))))...------------- ( -18.22, z-score = -2.16, R) >droEre2.scaffold_4820 6060151 92 - 10470090 ------AAAGUGUUAAAAUGCACUUAAAUAUUAAAGCCCG-CUUUCUGUGCCUAAGCCUCAAGUGAGCCCCGA--AUUAAUUCAAUUGCCAACACCCACCG------------- ------.((((((......))))))..........((.((-(((...(.((....)))..))))).)).....--..........................------------- ( -12.70, z-score = -1.17, R) >droYak2.chr3R 5973018 92 - 28832112 ------AAAGUGUUAAAAUGCACUUAAAUAUUAAAGCCCA-CUUUCUGGGCCUAAGCCCCAAGUGAGCCCUGA--AUUAAUUCAAUUGCCAGCACCCAUUG------------- ------.((((((......))))))..........((.((-(((...((((....)))).))))).))..(((--(....)))).................------------- ( -19.60, z-score = -1.86, R) >droSec1.super_22 264038 92 + 1066717 ------AAAGUGUUAAAAUGCACUUAAAUAUUAAAGCCCA-CUUUCUGGGCCUAAGCCCCAAGUGAGCCCUGA--AUUAAUUCAAUUGCCAACACCCACCG------------- ------.((((((......))))))..........((.((-(((...((((....)))).))))).))..(((--(....)))).................------------- ( -19.60, z-score = -2.71, R) >droSim1.chr3R 23351525 92 + 27517382 ------AAAGUGUUAAAAUGCACUUAAAUAUUAAAGCCCA-CUUUCUGGGCCUAAGCCCCAAGUGAGCCCUGA--AUUAAUUCAAUUGCCAACACCCACCG------------- ------.((((((......))))))..........((.((-(((...((((....)))).))))).))..(((--(....)))).................------------- ( -19.60, z-score = -2.71, R) >droMoj3.scaffold_6540 1912670 100 - 34148556 ------AAAGUGUUAAAAUGCACUUAAAUAUUAAAACUU----UGUCA--UCGUGCUCUCCAUCGGGAUUUG--CAUUAAUUCAAUUGCUUAGUUUUAUGAGAUUAAAUACGCA ------.((((((......))))))......((((((((----((...--..((((..(((....)))...)--))).....)))......)))))))................ ( -15.50, z-score = -0.12, R) >droVir3.scaffold_12855 8216191 96 - 10161210 ------AAAGUGUUAAAAUGCACUUAAAUAUUAAAACUUU-CGCAAUG---UCAGCUUUACAA-GGGACUUG--CAUUAAUUCAAUUGCUGAGUUUUAUGGGAUUAAGU----- ------.((((((......))))))......((((((((.-.(((((.---.........(((-(...))))--..........))))).))))))))...........----- ( -17.15, z-score = -0.37, R) >droWil1.scaffold_181089 3883828 91 + 12369635 -GCGAUAAAGUGUUAAAAAGCUCUUAAAUAUUAAAGCAUA-CAUUUUG-GUCUGAUAUUAUGUCAGACUUUGUUCAUUAAUUUAAUU--GCAUUUA------------------ -((((((((((((......(((.(((.....))))))..)-))))))(-(((((((.....))))))))...............)))--)).....------------------ ( -17.90, z-score = -1.78, R) >droGri2.scaffold_14830 5944804 107 - 6267026 UGAGCAAAAGUGUUAAAUUUCACUUAAAUACUAAAACUUUUCAUAUUGCAUGCGGCUCUCUUUUGGGAUUUG--CAUUAAUUUGAUUCAACAAUUUUAUGGGAUUAUAU----- ((((...(((((........)))))...............(((.((((.((((((.((((....)))).)))--))))))).)))))))....................----- ( -17.80, z-score = -0.81, R) >consensus ______AAAGUGUUAAAAUGCACUUAAAUAUUAAAGCCUA_CUUUCUGGGCCUAAGCCUCAAGUGAGCCCUGA__AUUAAUUCAAUUGCCAACACCCACCG_____________ .......((((((......))))))......................................................................................... ( -4.88 = -5.04 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:14 2011