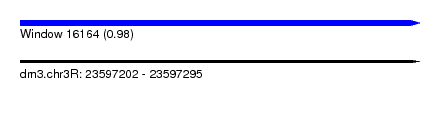
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,597,202 – 23,597,295 |
| Length | 93 |
| Max. P | 0.980688 |

| Location | 23,597,202 – 23,597,295 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 81.27 |
| Shannon entropy | 0.33537 |
| G+C content | 0.45950 |
| Mean single sequence MFE | -29.49 |
| Consensus MFE | -18.71 |
| Energy contribution | -19.17 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23597202 93 + 27905053 ---GCUGUCGAUUGCCACACUGUCACAAGUUGCGGGCGAAUAAAUCUUAUAAAUAUUUGAUGGCCUGCCAACCAGUCGGCAGCGACAU-AUCUAAAG- ---(((((((((((....(((......))).((((((.(.(((((.........))))).).))))))....))))))))))).....-........- ( -29.70, z-score = -3.09, R) >dp4.chr2 3953037 91 + 30794189 UCUUCUUUUGA--AUCGAGCUGCCAU---UUCUGGGCGGGCAAAACUUAUAAAUAUUUGAUAUCCUGCCUGGCAGAUGGUU-CGACAUUAUCUAAAG- ...........--.((((...(((((---(((..((((((((((...........))))....))))))..).))))))))-)))............- ( -24.50, z-score = -2.11, R) >droPer1.super_7 4045690 91 - 4445127 UCUUCUUUUGA--AUCGAGCUGCCAU---UUCUGGGCGGGCAAAACUUAUAAAUAUUUGAUAUCCUGCCUGGCAGAUGGUU-CGACAUUAUCUAAAG- ...........--.((((...(((((---(((..((((((((((...........))))....))))))..).))))))))-)))............- ( -24.50, z-score = -2.11, R) >droEre2.scaffold_4820 6027828 93 - 10470090 ---GCUGUCGAUUGCCGCACUGACACAAGUUGCGGGCGAAUACAUCUUAUAAAUAUUUGAUGGCCUGCCAGCCAGUCGGCAGCGACAU-AUCUAAGG- ---(((((((((((.((((((......)).))))((((....((((..((....))..))))...))))...))))))))))).....-........- ( -33.10, z-score = -2.75, R) >droYak2.chr3R 5941255 94 - 28832112 ---GCUGUCGAUAGCCGCACUGUCACAAGUUGCGGGCGAAUAAAUCUUAUAAAUAUUUGAUGGCCUGCCAGCCAGUCGGCAGCGACAU-AUCUAAAGC ---(((((((((..(((((((......)).))))).((((((...........)))))).((((......))))))))))))).....-......... ( -30.20, z-score = -1.89, R) >droSec1.super_22 234262 93 + 1066717 ---GCUGUCGAUUGCCGCACUGUCACAAGUUGCGGGCGAAUAAAUCUUAUAAAUAUUUGAUGGCCUGCCAACCAGUCGGCAGCGACAU-AUCUAAAG- ---..(((((.((((((.((((......(((((((((.(.(((((.........))))).).))))).))))))))))))))))))).-........- ( -32.20, z-score = -3.43, R) >droSim1.chr3R 23320634 93 + 27517382 ---GCUGUCGAUUGCCGCACUGUCACAAGUUGCGGGCGAAUAAAUCUUAUAAAUAUUUGAUGGCCUGCCAACCAGUCGGCAGCGACAU-AUCUAAAG- ---..(((((.((((((.((((......(((((((((.(.(((((.........))))).).))))).))))))))))))))))))).-........- ( -32.20, z-score = -3.43, R) >consensus ___GCUGUCGAUUGCCGCACUGUCACAAGUUGCGGGCGAAUAAAUCUUAUAAAUAUUUGAUGGCCUGCCAGCCAGUCGGCAGCGACAU_AUCUAAAG_ .....(((((...((((.((((......(((((((((.(.(((((.........))))).).))))).))))))))))))..)))))........... (-18.71 = -19.17 + 0.46)

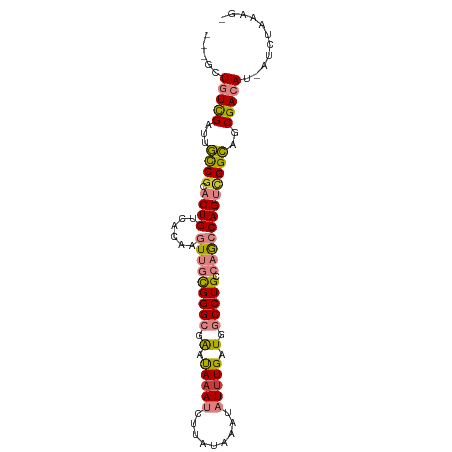
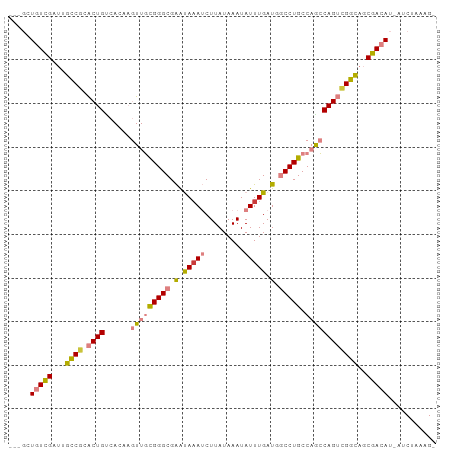
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:08 2011