| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,596,921 – 23,597,017 |
| Length | 96 |
| Max. P | 0.999760 |

| Location | 23,596,921 – 23,597,017 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.57 |
| Shannon entropy | 0.52390 |
| G+C content | 0.43844 |
| Mean single sequence MFE | -28.36 |
| Consensus MFE | -11.16 |
| Energy contribution | -11.15 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.848797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
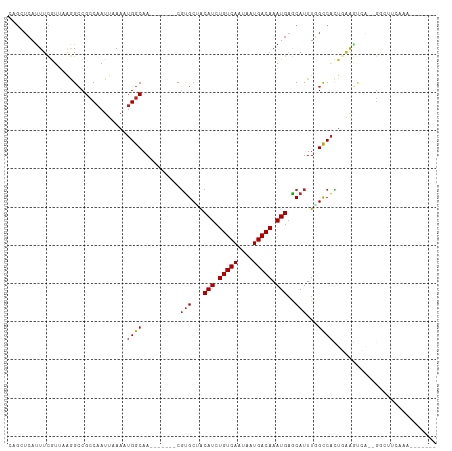
>dm3.chr3R 23596921 96 + 27905053 CAGAUCAUUUCGUUAAGGCCGCCAAUUAAAUUGGCAA-------CGUGCUGCAUCUGUCAAUAAUGACAAAUGAGCAUUUGGCCACUGGAGUCG--GGCUUCAAA------- ................((((((((((...))))))..-------.(((((.(((.(((((....))))).))))))))..))))..(((((...--..)))))..------- ( -32.10, z-score = -2.91, R) >droSim1.chr3R 23320355 96 + 27517382 CAGAUCAUUUCGUUAAGGCCGCCAAUUAAAUUGGCAA-------CGUGCUGCAUCUGUCAAUAAUGACAAAUGAGCAUUUGGCCACUGGAGUCG--GGCUUCAAA------- ................((((((((((...))))))..-------.(((((.(((.(((((....))))).))))))))..))))..(((((...--..)))))..------- ( -32.10, z-score = -2.91, R) >droSec1.super_22 233983 96 + 1066717 CAAAUCAUUUCGUUAAGGCCGCCAAUUAAAUUGGCAA-------CGUGCUGCAUCUGUCAAUAAUGACAAAUGAGCAUUUGGCCACUGGAGUCG--GGCUUCAAA------- ................((((((((((...))))))..-------.(((((.(((.(((((....))))).))))))))..))))..(((((...--..)))))..------- ( -32.10, z-score = -3.25, R) >droYak2.chr3R 5940989 96 - 28832112 CAGAUCAUUUCGUUAAGGCCGCCAAUUAAAAUGGCAA-------CGUGCUGCAUGUGUCAAUAAUGACAAAUGAGCAUUUGGCCACUGAAGUCG--GGCUUCAAA------- ................((((((((.......))))..-------.(((((.(((.(((((....))))).))))))))..))))..(((((...--..)))))..------- ( -31.20, z-score = -2.68, R) >droEre2.scaffold_4820 6027590 96 - 10470090 CAGAUCAUUUCGUUAAGGCCGCCAAUUAAAAUGGCAA-------CGUGCUGCAUCUGUCAAUAAUGACAAAUGAGCAUUUGGCCACUGAAGUCG--GGCUUCAAA------- ................((((((((.......))))..-------.(((((.(((.(((((....))))).))))))))..))))..(((((...--..)))))..------- ( -31.20, z-score = -3.01, R) >droAna3.scaffold_13340 4605338 105 - 23697760 CGGCUCAUUUCGUAAAGGGCGCCAAUUAAAAAGGCAA-------CCUGCAACAUCUGUCAAUAAUGACAAAUGAGCAUUUGGUCGGACAACCGAUCGGUGGAAGCCUUCAAA ..((((((((.((..(((..(((.........)))..-------)))...))....((((....))))))))))))....((((((....))))))(((....)))...... ( -32.40, z-score = -2.70, R) >droPer1.super_7 4045401 112 - 4445127 CAGCUCAUUUCAUUUAAGCCGCCAAUUAAAAUGGCAACGGGCAACGUGCAACAUCUGUCAAUAAUGACAAAUGAGCAUUUGGCCAUUCAGGCGAUCGAAGCGGCGACUCAAA ..(((..((((......(((((((.......))))...((.(((.((((..(((.(((((....))))).))).))))))).)).....)))....)))).)))........ ( -31.00, z-score = -1.62, R) >dp4.chr2 3952756 112 + 30794189 CAGCUCAUUUCAUUUAAGCCGCCAAUUAAAAUGGCAACGGGCAACGUGCAACAUCUGUCAAUAAUGACAAAUGAGCAUUUGGCCAUUCAGGCGAUCGAAGCGGCGACUCAAA ..(((..((((......(((((((.......))))...((.(((.((((..(((.(((((....))))).))).))))))).)).....)))....)))).)))........ ( -31.00, z-score = -1.62, R) >droWil1.scaffold_181089 3845602 85 + 12369635 CAGCUCAUUUCAUUAAGGCCGGCAAUUAAAAUGGCAA-------CGCGCCGCAUCUGUCAAUAAUGACAAAUGUGCAUUUCGCCAAUCCAAA-------------------- ................((..(((.........(((..-------...)))((((.(((((....)))))...)))).....)))...))...-------------------- ( -19.60, z-score = -0.96, R) >droVir3.scaffold_12855 8176832 96 - 10161210 -UAUGGACUUUGUUUGAGCCGCCAAUUAAAAUGGCAA------ACGUGCGACAUCUGUCAAUAAUGACAAAUGAACAUUUCGCCAUUAAAGCCA--GGCCCAGAA------- -...........((((.(((((((.......))))..------..(.(((((((.(((((....))))).)))......)))))..........--))).)))).------- ( -23.20, z-score = -1.50, R) >droMoj3.scaffold_6540 1869334 87 - 34148556 CUGCUCAUUUUGUGUAAGCCGCCAAUUAAAAUGGCAA------ACGUGCGACAUCUGUCAAUAAUGACAAAUGAACAUUUUGCCAUUAAAGCC------------------- ..(((......(((.....))).......((((((((------(.(((...(((.(((((....))))).)))..))))))))))))..))).------------------- ( -19.90, z-score = -1.64, R) >droGri2.scaffold_14830 5906113 104 - 6267026 CAGCUCAUUUUGUGUAAGCCGCCAAUUAAAAUGGCAA------ACGUGCGACAUCUGUCAAUAAUGACAAAUGAACAUUUCGCCAUUAAGCUCA--GCAGCCGGGCCAAAGU ..((((...((((...(((.((((.......))))..------..(.(((((((.(((((....))))).)))......))))).....)))..--))))..))))...... ( -24.50, z-score = -0.59, R) >consensus CAGCUCAUUUCGUUAAGGCCGCCAAUUAAAAUGGCAA_______CGUGCUACAUCUGUCAAUAAUGACAAAUGAGCAUUUGGCCACUGAAGUCA__GGCUUCAAA_______ .................((.((((.......))))..........((....(((.(((((....))))).))).)).....))............................. (-11.16 = -11.15 + -0.01)


| Location | 23,596,921 – 23,597,017 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.57 |
| Shannon entropy | 0.52390 |
| G+C content | 0.43844 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -19.67 |
| Energy contribution | -19.58 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.33 |
| SVM RNA-class probability | 0.999760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
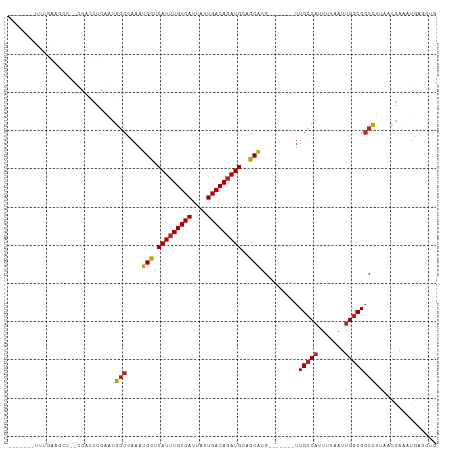
>dm3.chr3R 23596921 96 - 27905053 -------UUUGAAGCC--CGACUCCAGUGGCCAAAUGCUCAUUUGUCAUUAUUGACAGAUGCAGCACG-------UUGCCAAUUUAAUUGGCGGCCUUAACGAAAUGAUCUG -------.........--..........((((...(((((((((((((....))))))))).))))..-------..((((((...))))))))))................ ( -30.80, z-score = -3.22, R) >droSim1.chr3R 23320355 96 - 27517382 -------UUUGAAGCC--CGACUCCAGUGGCCAAAUGCUCAUUUGUCAUUAUUGACAGAUGCAGCACG-------UUGCCAAUUUAAUUGGCGGCCUUAACGAAAUGAUCUG -------.........--..........((((...(((((((((((((....))))))))).))))..-------..((((((...))))))))))................ ( -30.80, z-score = -3.22, R) >droSec1.super_22 233983 96 - 1066717 -------UUUGAAGCC--CGACUCCAGUGGCCAAAUGCUCAUUUGUCAUUAUUGACAGAUGCAGCACG-------UUGCCAAUUUAAUUGGCGGCCUUAACGAAAUGAUUUG -------.........--..........((((...(((((((((((((....))))))))).))))..-------..((((((...))))))))))................ ( -30.80, z-score = -3.30, R) >droYak2.chr3R 5940989 96 + 28832112 -------UUUGAAGCC--CGACUUCAGUGGCCAAAUGCUCAUUUGUCAUUAUUGACACAUGCAGCACG-------UUGCCAUUUUAAUUGGCGGCCUUAACGAAAUGAUCUG -------.((((((..--...)))))).((((...(((((((.(((((....))))).))).))))..-------..((((.......))))))))................ ( -30.50, z-score = -3.28, R) >droEre2.scaffold_4820 6027590 96 + 10470090 -------UUUGAAGCC--CGACUUCAGUGGCCAAAUGCUCAUUUGUCAUUAUUGACAGAUGCAGCACG-------UUGCCAUUUUAAUUGGCGGCCUUAACGAAAUGAUCUG -------.((((((..--...)))))).((((...(((((((((((((....))))))))).))))..-------..((((.......))))))))................ ( -34.10, z-score = -4.30, R) >droAna3.scaffold_13340 4605338 105 + 23697760 UUUGAAGGCUUCCACCGAUCGGUUGUCCGACCAAAUGCUCAUUUGUCAUUAUUGACAGAUGUUGCAGG-------UUGCCUUUUUAAUUGGCGCCCUUUACGAAAUGAGCCG ......(((((....((...((((....))))...(((.(((((((((....)))))))))..)))((-------.((((.........)))).))....))....))))). ( -34.00, z-score = -3.42, R) >droPer1.super_7 4045401 112 + 4445127 UUUGAGUCGCCGCUUCGAUCGCCUGAAUGGCCAAAUGCUCAUUUGUCAUUAUUGACAGAUGUUGCACGUUGCCCGUUGCCAUUUUAAUUGGCGGCUUAAAUGAAAUGAGCUG (((((((((((((.......))..(((((((.....((.(((((((((....)))))))))..))(((.....))).))))))).....)))))))))))............ ( -37.80, z-score = -3.12, R) >dp4.chr2 3952756 112 - 30794189 UUUGAGUCGCCGCUUCGAUCGCCUGAAUGGCCAAAUGCUCAUUUGUCAUUAUUGACAGAUGUUGCACGUUGCCCGUUGCCAUUUUAAUUGGCGGCUUAAAUGAAAUGAGCUG (((((((((((((.......))..(((((((.....((.(((((((((....)))))))))..))(((.....))).))))))).....)))))))))))............ ( -37.80, z-score = -3.12, R) >droWil1.scaffold_181089 3845602 85 - 12369635 --------------------UUUGGAUUGGCGAAAUGCACAUUUGUCAUUAUUGACAGAUGCGGCGCG-------UUGCCAUUUUAAUUGCCGGCCUUAAUGAAAUGAGCUG --------------------.(((((.((((((..(((.(((((((((....)))))))))..)))..-------)))))).)))))....((((.(((......))))))) ( -28.30, z-score = -2.57, R) >droVir3.scaffold_12855 8176832 96 + 10161210 -------UUCUGGGCC--UGGCUUUAAUGGCGAAAUGUUCAUUUGUCAUUAUUGACAGAUGUCGCACGU------UUGCCAUUUUAAUUGGCGGCUCAAACAAAGUCCAUA- -------...((((((--..(((..(((((((((.(((.(((((((((....)))))))))..)))..)------))))))))......))))))))).............- ( -34.00, z-score = -3.98, R) >droMoj3.scaffold_6540 1869334 87 + 34148556 -------------------GGCUUUAAUGGCAAAAUGUUCAUUUGUCAUUAUUGACAGAUGUCGCACGU------UUGCCAUUUUAAUUGGCGGCUUACACAAAAUGAGCAG -------------------.(((..(((((((((.(((.(((((((((....)))))))))..)))..)------))))))))......))).(((((.......))))).. ( -29.70, z-score = -4.16, R) >droGri2.scaffold_14830 5906113 104 + 6267026 ACUUUGGCCCGGCUGC--UGAGCUUAAUGGCGAAAUGUUCAUUUGUCAUUAUUGACAGAUGUCGCACGU------UUGCCAUUUUAAUUGGCGGCUUACACAAAAUGAGCUG ..((((....((((((--..(....(((((((((.(((.(((((((((....)))))))))..)))..)------))))))))....)..))))))....))))........ ( -39.20, z-score = -4.38, R) >consensus _______UUUGAAGCC__CGACUUCAAUGGCCAAAUGCUCAUUUGUCAUUAUUGACAGAUGCAGCACG_______UUGCCAUUUUAAUUGGCGGCCUUAACGAAAUGAGCUG ............................(((....(((.(((((((((....)))))))))..)))..........(((((.......))))))))................ (-19.67 = -19.58 + -0.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:07 2011