
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,578,925 – 23,579,034 |
| Length | 109 |
| Max. P | 0.719785 |

| Location | 23,578,925 – 23,579,034 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 88.74 |
| Shannon entropy | 0.21383 |
| G+C content | 0.48535 |
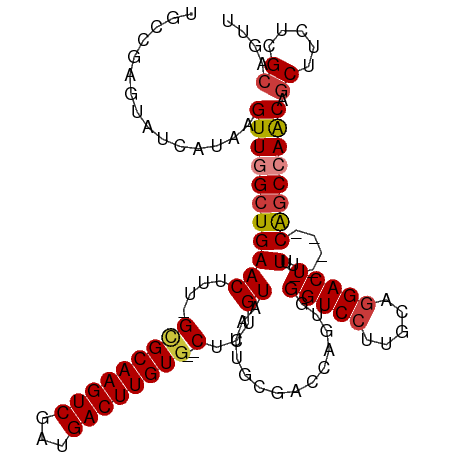
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -24.86 |
| Energy contribution | -25.37 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.719785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
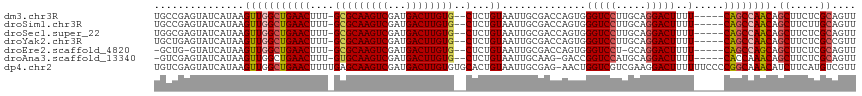
>dm3.chr3R 23578925 109 + 27905053 UGCCGAGUAUCAUAAGUUGGCUGAACUUU-GCGCAAGUCGAUGACUUGUG--CUCUGUAAUUGCGACCAGUGGGUCCUUGCAGGACUUUU-----CAGCCAACAGCUUCUCGCAGUU (((.(((........((((((((((....-(((((((((...))))))))--)(((((((..(.((((....))))))))))))....))-----)))))))).....))))))... ( -43.22, z-score = -3.47, R) >droSim1.chr3R 23302286 109 + 27517382 UGCCGAGUAUCAUAAGUUGGCUGAACUUU-GCGCAAGUCGAUGACUUGUG--CUCUGUAAUUGCGACCAGUGGGUCCUUGCAGGACUUUU-----CAGCCAACAGCUUCUUGCAGUU ...............((((((((((....-(((((((((...))))))))--)(((((((..(.((((....))))))))))))....))-----)))))))).((.....)).... ( -41.50, z-score = -2.99, R) >droSec1.super_22 215963 109 + 1066717 UGGCGAGUAUCAUAAGUUGGCUGAACUUU-GCGCAAGUCGAUGACUUGUG--CUCUGUAAUUGCGACCAGUGGGUCCUUGCAGGACUUUU-----CAGCCAACAGCUUCUCGCAGUU ..(((((........((((((((((....-(((((((((...))))))))--)(((((((..(.((((....))))))))))))....))-----)))))))).....))))).... ( -46.32, z-score = -4.13, R) >droYak2.chr3R 5921740 109 - 28832112 UGCUGAGUAUCAUAAGUUGGCUGAACUUU-GCGCAAGUCGAUGACUUGUG--CUCUGUAAUUGCGACCAGUGGGUCCUUGCAGGACUUUU-----CAGCCAACAGCUUCUCGCCGUU .((.(((........((((((((((....-(((((((((...))))))))--)(((((((..(.((((....))))))))))))....))-----)))))))).....))))).... ( -41.62, z-score = -3.08, R) >droEre2.scaffold_4820 6010573 106 - 10470090 -GCUG-GUAUCAUAAGUUGGCUGAACUUU-GCGCAAGUCGAUGACUUGUG--CUCUGUAAUUGCGACCAGUGGGUCCU-GCAGGACUUUU-----CAGCCAGCAGCUUCUCGCAGUU -((((-.........((((((((((....-(((((((((...))))))))--)((((((...(.((((....))))))-)))))....))-----)))))))).........)))). ( -39.77, z-score = -2.07, R) >droAna3.scaffold_13340 4583394 107 - 23697760 -GUCGAGUAUCAUAAGUUGGCUGAACUUU-GUGCAAGUCGAUGACUUGUG--CUCUGUAAUUGCAAG-GACCGGUCCAUGCAGGACUUUU-----CACCAAACAGCUUCUCGCAGUU -(.((((......((((((..((.((...-(..((((((...))))))..--)...))..(((((.(-((....))).))))).......-----...))..))))))))))).... ( -30.70, z-score = -0.84, R) >dp4.chr2 3933841 116 + 30794189 UGUCGAGUAUCAUAAGUUGGCUGAACUUUUGAGCAAGUCGAUGACUUGUGUGCACUGUAAUUGCGAG-AACUGGUCGUCGAAGGACUUUUUUCCCCGGCAAACAUCUUCAUGUCGUU (((((.((.(((.(((((.....))))).)))))...(((((((((.((.((((.......))))..-.)).))))))))).(((......))).)))))................. ( -29.70, z-score = -0.01, R) >consensus UGCCGAGUAUCAUAAGUUGGCUGAACUUU_GCGCAAGUCGAUGACUUGUG__CUCUGUAAUUGCGACCAGUGGGUCCUUGCAGGACUUUU_____CAGCCAACAGCUUCUCGCAGUU ...............((((((((.((.....((((((((...))))))))......))..............(((((.....)))))........)))))))).((.....)).... (-24.86 = -25.37 + 0.51)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:04 2011