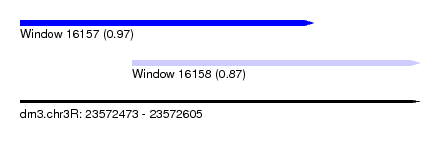
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,572,473 – 23,572,605 |
| Length | 132 |
| Max. P | 0.973140 |

| Location | 23,572,473 – 23,572,570 |
|---|---|
| Length | 97 |
| Sequences | 14 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 67.36 |
| Shannon entropy | 0.65581 |
| G+C content | 0.63288 |
| Mean single sequence MFE | -39.30 |
| Consensus MFE | -21.26 |
| Energy contribution | -20.87 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.973140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
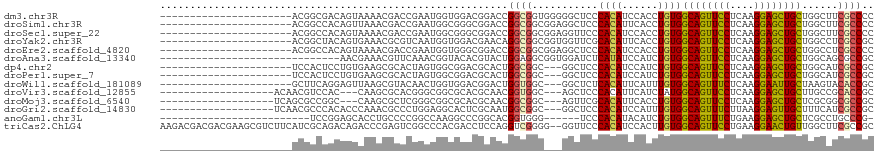
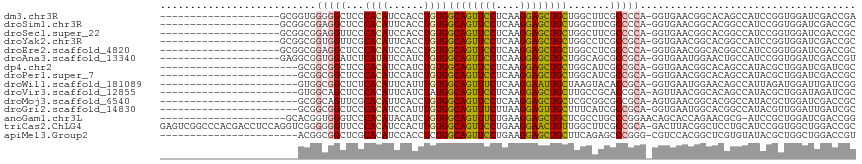
>dm3.chr3R 23572473 97 + 27905053 ----------------------ACGGCGACAGUAAAACGACCGAAUGGUGGACGGACCGGCGGUGGGGGCUCCCACAUCCACCUGUGGCAGUUCCUCAAGGAGCUGCUGGCUUCGCCCC ----------------------..(((((.(((.............((((((((......))(((((....))))).))))))...(((((((((....))))))))).)))))))).. ( -43.90, z-score = -1.71, R) >droSim1.chr3R 23295902 97 + 27517382 ----------------------ACGGCCACAGUUAAACGACCGAAUGGCGGGCGGACCGGCGGCGGAGGCUCCCACAUUCACCUGUGGCAGUUCCUCAAGGAGCUGCUGGCUUCGCCCC ----------------------...(((((.(((....))).)..))))(((((((((((((((.((((((.(((((......))))).))..)))).....)))))))).))))))). ( -45.60, z-score = -2.25, R) >droSec1.super_22 209615 97 + 1066717 ----------------------ACGGCCACAGUAAAACGACCGAAUGGCGGGCGGACCGGCGGCGGAGGUUCCCACAUCCACCUGUGGCAGUUCCUCAAGGAGCUGCUGGCUUCGCCCC ----------------------...(((((.((......)).)..))))(((((((((((((((.((((..((((((......)))))..)..)))).....)))))))).))))))). ( -44.30, z-score = -1.80, R) >droYak2.chr3R 5914975 97 - 28832112 ----------------------ACGGCUACAGUGAAACGCGUCAAUGGUGGACGAACAGGCGGCGGUGGUUCGCACAUUCACCUGUGGCAGUUCCUCAAGGAGCUGCUGGCCUCGCCGC ----------------------.((.((((..(((......)))...)))).)).....(((((((.((((..((((......))))((((((((....)))))))).))))))))))) ( -40.20, z-score = -1.23, R) >droEre2.scaffold_4820 6004397 97 - 10470090 ----------------------ACGGCCACAGUAAAACGACCGAAUGGUGGGCGGACCGGCGGCGGAGGCUCCCACAUCCACCUGUGGCAGUUCCUCAAGGAGCUGCUGGCCUCGCCCC ----------------------.((.((((.................)))).))....((.((((.(((((..((((......))))((((((((....)))))))).))))))))))) ( -41.73, z-score = -0.83, R) >droAna3.scaffold_13340 4576687 89 - 23697760 ------------------------------AACGAAACGUUCAAACGGUACACGUACUGGAGGCGGUGGAUCUCAUAUCCAUCUGUGGCAGUUCCUCAAGGAGCUGCUGGCAGCGCCGC ------------------------------...............(((((....)))))..((((((((((.....))))))(((((((((((((....))))))))).)))))))).. ( -35.00, z-score = -2.63, R) >dp4.chr2 3926036 94 + 30794189 ----------------------UCCACUCCUGUGAAGCGCACUAGUGGCGGACGCACUGGCGGC---GGCUCCCACAUCCAUCUGUGGCAGUUCCUCAAGGAGCUGCUGGCAUCGCCGC ----------------------.(((((..((((...))))..)))))(((.....)))(((((---((..((((((......))))((((((((....)))))))).))..))))))) ( -39.60, z-score = -1.14, R) >droPer1.super_7 4020622 94 - 4445127 ----------------------UCCACUCCUGUGAAGCGCACUAGUGGCGGACGCACUGGCGGC---GGCUCCCACAUCCAUCUGUGGCAGUUCCUCAAGGAGCUGCUGGCAUCGCCGC ----------------------.(((((..((((...))))..)))))(((.....)))(((((---((..((((((......))))((((((((....)))))))).))..))))))) ( -39.60, z-score = -1.14, R) >droWil1.scaffold_181089 3814265 94 + 12369635 ----------------------GCUUCAGGAGUUAAGCGUACAACUGGUGGACGGACUGGUGGC---GGCUCUCACAUUCAUUUGUGGCAGUUUCUCAAGGAAUUGCUAAGUACACCGC ----------------------((((........))))......(((.....)))...((((..---.(((..((((......))))((((((((....))))))))..))).)))).. ( -24.40, z-score = 0.32, R) >droVir3.scaffold_12855 8137946 94 - 10161210 -------------------ACAACGUCCAC---CAAGCGCACGGGCGGCGCACGCAACGGUGGC---AGCUCCCACAUUCAUCUAUGGCAGUUCCUCAAGGAGCUGCUUGCCGCACCGC -------------------....(((....---...)))..((((((((((.(((....)))))---...................(((((((((....))))))))).))))).))). ( -34.80, z-score = -1.19, R) >droMoj3.scaffold_6540 1833178 94 - 34148556 -------------------UCAGCGCCGGC---CAAGCGCUCGGGCGGCGCACGCAACGGCGGC---AGUUCGCACAUUCACCUGUGGCAGUUCCUCAAGGAGCUGCUCGCGGCGCCGC -------------------...((((((.(---(........)).))))))......(((((((---.....))........(((((((((((((....)))))))).)))))))))). ( -48.50, z-score = -2.11, R) >droGri2.scaffold_14830 5871145 97 - 6267026 -------------------UCAACGCCCACACCCAAACGCCCUGGAGGCACUCGCAAUGGCGGC---GGCUCCCACAUCCAUUUGUGGCAGUUUCUUAAGGAGUUGCUUUCAUCGCCGC -------------------....((((.....(((.......)))..((....))...))))((---(((...((((......))))(((..(((....)))..))).......))))) ( -28.30, z-score = -0.05, R) >anoGam1.chr3L 20691624 87 - 41284009 -------------------------UCCGGAGCACCUGCCCCGGCCAAGGCCCGGCACGGUGGG------UCCCACAUACAUCUGUGGCAGUUUCUGAAGGAGCUGCUCGCCUGCCCG- -------------------------..(((.(((((((((..(((....))).)))).)))(((------(..((((......))))((((((((....))))))))..)))))))))- ( -37.90, z-score = -1.16, R) >triCas2.ChLG4 8221446 117 + 13894384 AAGACGACGACGAAGCGUCUUCAUCGCAGACAGACCCGAGUCGGCCCACGACCUCCAGGUCGGGG--GGUUCCCACAUCCACUUGUGGCAGUUCCUGAAGGAACUGUUGGCUUCGCCGC .......((.((((((((((.......)))).(((....)))(((((.(((((....)))))..)--)))).(((((......)))))(((((((....)))))))...)))))).)). ( -46.30, z-score = -1.32, R) >consensus ______________________ACGGCCACAGUAAAACGCCCGAAUGGCGGACGCACCGGCGGC___GGCUCCCACAUCCACCUGUGGCAGUUCCUCAAGGAGCUGCUGGCUUCGCCGC ..........................................................((((...........((((......))))((((((((....))))))))......)))).. (-21.26 = -20.87 + -0.38)
| Location | 23,572,510 – 23,572,605 |
|---|---|
| Length | 95 |
| Sequences | 15 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.78 |
| Shannon entropy | 0.46629 |
| G+C content | 0.62972 |
| Mean single sequence MFE | -41.63 |
| Consensus MFE | -20.36 |
| Energy contribution | -20.41 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
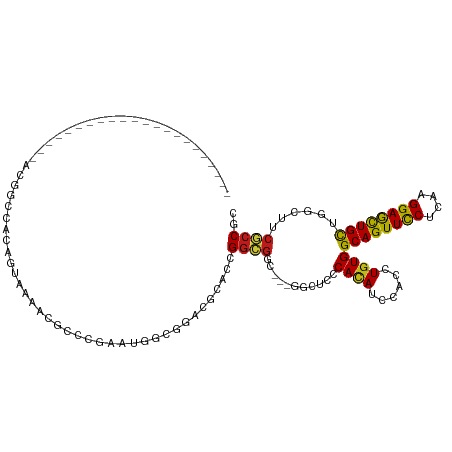
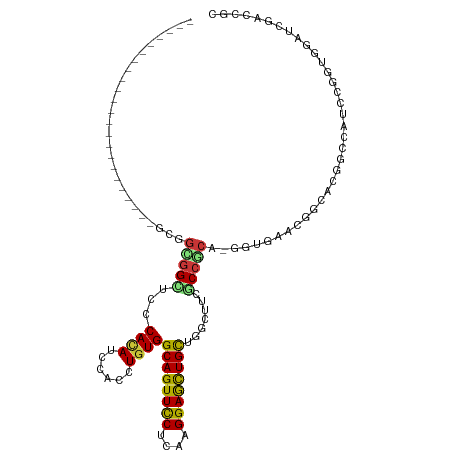
>dm3.chr3R 23572510 95 + 27905053 --------------------GCGGUGGGGGCUCCCACAUCCACCUGUGGCAGUUCCUCAAGGAGCUGCUGGCUUCGCCCCA-GGUGAACGGCACAGCCAUCCGGUGGAUCGACCGA --------------------.((((.(((....))..(((((((.(((((((((((....))))))((((...(((((...-))))).))))...)))))..)))))))).)))). ( -45.80, z-score = -1.80, R) >droSim1.chr3R 23295939 95 + 27517382 --------------------GCGGCGGAGGCUCCCACAUUCACCUGUGGCAGUUCCUCAAGGAGCUGCUGGCUUCGCCCCA-GGUGAACGGCACGGCCAUCCGGUGGAUCGACCGC --------------------(((((((.((((....(.((((((((.(((((((((....)))))))))(((...))).))-)))))).)....)))).(((...)))))).)))) ( -44.90, z-score = -1.86, R) >droSec1.super_22 209652 95 + 1066717 --------------------GCGGCGGAGGUUCCCACAUCCACCUGUGGCAGUUCCUCAAGGAGCUGCUGGCUUCGCCCCA-GGUGAACGGCACGGCCAUCCGGUGGAUCGACCGC --------------------(((((((.((...))...((((((.(((((((((((....))))))((((...(((((...-))))).))))...)))))..))))))))).)))) ( -43.80, z-score = -1.57, R) >droYak2.chr3R 5915012 95 - 28832112 --------------------GCGGCGGUGGUUCGCACAUUCACCUGUGGCAGUUCCUCAAGGAGCUGCUGGCCUCGCCGCA-GGUGAACGGCACGGCCAUCCGGUGGAUCGACCGC --------------------...((((((((((((...((((((((((((((((((....)))))))))(((...))))))-)))))).(((...))).....))))))).))))) ( -50.80, z-score = -3.27, R) >droEre2.scaffold_4820 6004434 95 - 10470090 --------------------GCGGCGGAGGCUCCCACAUCCACCUGUGGCAGUUCCUCAAGGAGCUGCUGGCCUCGCCCCA-GGUGAACGGCACGGCCAUCCGGUGGAUCGACCGC --------------------(((((((.((...))...((((((...(((((((((....)))))))))((((..(((...-.......)))..))))....))))))))).)))) ( -44.80, z-score = -1.43, R) >droAna3.scaffold_13340 4576716 95 - 23697760 --------------------GAGGCGGUGGAUCUCAUAUCCAUCUGUGGCAGUUCCUCAAGGAGCUGCUGGCAGCGCCGCA-GGUGAAUGGAACUGCCAUCCGGUGGAUCGACCGU --------------------...(((((.((((.(((...((((((((((((((((....))))))((.....))))))))-)))).((((.....))))...))))))).))))) ( -45.60, z-score = -3.08, R) >dp4.chr2 3926073 92 + 30794189 -----------------------GCGGCGGCUCCCACAUCCAUCUGUGGCAGUUCCUCAAGGAGCUGCUGGCAUCGCCGCA-GGUGAACGGCACAGCCAUACGCUGGAUCGAUCGC -----------------------(((((((..((((((......))))((((((((....)))))))).))..))))))).-.((((.(((..((((.....))))..))).)))) ( -44.60, z-score = -3.20, R) >droPer1.super_7 4020659 92 - 4445127 -----------------------GCGGCGGCUCCCACAUCCAUCUGUGGCAGUUCCUCAAGGAGCUGCUGGCAUCGCCGCA-GGUGAACGGCACAGCCAUACGCUGGAUCGACCGC -----------------------(((((((..((((((......))))((((((((....)))))))).))..))))))).-(((...(((..((((.....))))..)))))).. ( -43.10, z-score = -2.64, R) >droWil1.scaffold_181089 3814302 92 + 12369635 -----------------------GUGGCGGCUCUCACAUUCAUUUGUGGCAGUUUCUCAAGGAAUUGCUAAGUACACCGCA-GGUGAAUGGAACAGCCAUUAGAUGGAUUGAUCGG -----------------------.....((((.(..((((((((((((((((((((....))))))))...(....)))))-))))))))..).))))....(((......))).. ( -30.00, z-score = -2.17, R) >droVir3.scaffold_12855 8137983 92 - 10161210 -----------------------GUGGCAGCUCCCACAUUCAUCUAUGGCAGUUCCUCAAGGAGCUGCUUGCCGCACCGCA-AGUUAACGGCACAGCCAUACGCUGGAUAGAUCGC -----------------------((((......))))....(((((((((((((((....)))))))))(((((...(...-.)....)))))((((.....)))).))))))... ( -35.50, z-score = -2.91, R) >droMoj3.scaffold_6540 1833215 92 - 34148556 -----------------------GCGGCAGUUCGCACAUUCACCUGUGGCAGUUCCUCAAGGAGCUGCUCGCGGCGCCGCA-AGUGAACGGCACGGCCAUACGCUGGAUCGACCGC -----------------------((((..(.((((.(.(((((.((((((((((((....))))))((.....))))))))-.))))).))).((((.....)))))).)..)))) ( -41.20, z-score = -1.99, R) >droGri2.scaffold_14830 5871185 92 - 6267026 -----------------------GCGGCGGCUCCCACAUCCAUUUGUGGCAGUUUCUUAAGGAGUUGCUUUCAUCGCCGCA-GGUGAAUGGCACGGCCAUACGUUGGAUUGAUCGC -----------------------(((((((.(((.((...(((((((((((((..(((...)))..)))......))))))-)))).(((((...)))))..)).)))))).)))) ( -29.70, z-score = -0.32, R) >anoGam1.chr3L 20691653 94 - 41284009 ---------------------GCACGGUGGGUCCCACAUACAUCUGUGGCAGUUUCUGAAGGAGCUGCUCGCCUGCCCGGAACAGCACCAGAACGCG-AUCCGCUGGAUCGACCGG ---------------------(((.((((((..(((((......)))))(((((((....))))))))))))))))((((....((........))(-((((...)))))..)))) ( -36.70, z-score = -1.34, R) >triCas2.ChLG4 8221483 115 + 13894384 GAGUCGGCCCACGACCUCCAGGUCGGGGGGUUCCCACAUCCACUUGUGGCAGUUCCUGAAGGAACUGUUGGCUUCGCCGCA-GACUUACGGCUCCUGCAUCCGGUGGCUGGACCGC ((((((((((.(((((....)))))..))))).(((((......)))))(((((((....)))))))..(((...)))...-))))).(((.(((.((........)).)))))). ( -48.40, z-score = -0.86, R) >apiMel3.Group2 10266711 92 + 14465785 -----------------------ACGGCGGCUCGCACAUCCACCUCUGGCAGUUCCUGAAGGAGCUGCUUCAGAGCCCGGG-CGUCCACGGCUCGUGUAUACGCUGGCUGGACCGU -----------------------(((((((..(((....((..(((((((((((((....))))))))..)))))...)))-)).)).(((((.(((....))).)))))).)))) ( -39.60, z-score = -1.08, R) >consensus _______________________GCGGCGGCUCCCACAUCCACCUGUGGCAGUUCCUCAAGGAGCUGCUGGCUUCGCCGCA_GGUGAACGGCACGGCCAUCCGGUGGAUCGACCGC ..........................(((((...((((......))))((((((((....)))))))).......))))).................................... (-20.36 = -20.41 + 0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:49:03 2011