
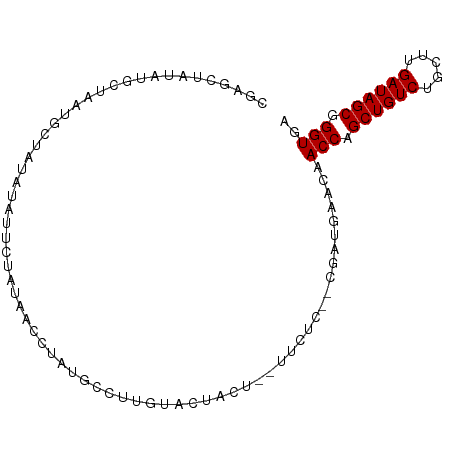
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,492,437 – 23,492,536 |
| Length | 99 |
| Max. P | 0.950814 |

| Location | 23,492,437 – 23,492,528 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 78.48 |
| Shannon entropy | 0.43992 |
| G+C content | 0.39577 |
| Mean single sequence MFE | -16.89 |
| Consensus MFE | -12.17 |
| Energy contribution | -12.17 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.616739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23492437 91 + 27905053 CGAGCUAUAUGCUAAUGCUAUAUCUUCUAUAACCUAUGCCUUGUACUACU--UUCUC--CGAUGAACAACCAGCUGUCUGCUUGAUAGCGGGUGA .(((.((((.((....)))))).))).....((((...............--(((..--....)))......((((((.....)))))))))).. ( -16.70, z-score = -0.21, R) >droSim1.chr3R 23222939 91 + 27517382 CGUGCUAUAUGCUAAUGCUAUAUCUUCUAUAACCCAUGCCUUGUACUACU--UUCUC--CGAUGAACAACCAGCUGUCUGCUUGAUAGCGGGUGA .....((((.((....)))))).........((((...............--(((..--....)))......((((((.....)))))))))).. ( -17.40, z-score = -0.55, R) >droSec1.super_22 135679 91 + 1066717 CGUGCUAUAUGCUAAUGCUAUAUCUUCUAUAACCUAUGCCUUGUACUACU--UUCUC--CGAUGAACAACCAGCUGUCUGCUUGAUAGCGGGUGA .((((.....((....(.((((.....)))).)....))...))))....--(((..--....)))..(((.((((((.....)))))).))).. ( -16.10, z-score = -0.15, R) >droYak2.chr3R 5837405 92 - 28832112 CGAGCUAUUUGCUAAUGCUAUAUAUUCUAUACCCUAUGCCUUGUACUACUC-UUCUC--CAAUGAACAACCAGCUGUCUGCUUGAUAGCGGGUGA .(((.(((..((....))...))))))..(((((.................-(((..--....)))......((((((.....))))))))))). ( -18.60, z-score = -1.06, R) >droEre2.scaffold_4820 5925106 85 - 10470090 CGAGCUAUU-GCUAAUGCUAUAUAUUCUAUACCCUAUG-----UACUACU--UUCUC--CAAUGAACAACCAGCUGUCUGCUUGAUAGCGGGUGA ..(((....-)))................(((((....-----.......--(((..--....)))......((((((.....))))))))))). ( -18.50, z-score = -1.48, R) >droAna3.scaffold_13340 4502077 86 - 23697760 CAAGCUA-AUACU----UUAUGUAUUCUACCACUUUUGACUUGUAAAUAU--UUCUU--CCAUGAUCAACCAGCUGUCUGCUCGAUAGCGGGUGA ((((..(-((((.----....)))))....((....)).)))).......--.....--.........(((.((((((.....)))))).))).. ( -13.50, z-score = -0.00, R) >dp4.chr2 3847679 85 + 30794189 UAAGCUA-AUGCUA---CUAUAUAUUCUAUACCUUAUG--UACUACAACU--UUCUC--UUAUGAUCAACCAGCUGUCUGCUUGAUAGCGGGUGA ..(((..-..))).---.((((((..........))))--))........--.....--.........(((.((((((.....)))))).))).. ( -15.50, z-score = -0.86, R) >droPer1.super_7 3941903 85 - 4445127 UAAGCUA-AUGCUA---CUAUAUAUUCUAUACCUUAUG--UACUACAACU--UUCUC--UUAUGAUCAACCAGCUGUCUGCUUGAUAGCGGGUGA ..(((..-..))).---.((((((..........))))--))........--.....--.........(((.((((((.....)))))).))).. ( -15.50, z-score = -0.86, R) >droWil1.scaffold_181089 3733824 79 + 12369635 ----------------UAUAUAUAUAUUACUAUGUAUAUAUUCUAUGAUUCAUUCUUUACUAUGAACAACCAGCUGUCUGCUCGAUAGCGGGUGA ----------------((((((((((....)))))))))).....((.(((((........)))))))(((.((((((.....)))))).))).. ( -20.20, z-score = -2.94, R) >consensus CGAGCUAUAUGCUAAUGCUAUAUAUUCUAUAACCUAUGCCUUGUACUACU__UUCUC__CGAUGAACAACCAGCUGUCUGCUUGAUAGCGGGUGA ....................................................................(((.((((((.....)))))).))).. (-12.17 = -12.17 + -0.00)


| Location | 23,492,445 – 23,492,536 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 72.15 |
| Shannon entropy | 0.58967 |
| G+C content | 0.39240 |
| Mean single sequence MFE | -17.45 |
| Consensus MFE | -12.20 |
| Energy contribution | -12.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.950814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23492445 91 + 27905053 AUGCUAAUGCUAUAUCUUCUAUAACCUAUGCCUUGUACUACU-UUCUCCGAUGAACAACCAGCUGUCUGCUUGAUAGCGGGUGAGUUUCGGU ..((....))................................-....((((..(((.(((.((((((.....)))))).)))..))))))). ( -19.10, z-score = -0.70, R) >droSim1.chr3R 23222947 91 + 27517382 AUGCUAAUGCUAUAUCUUCUAUAACCCAUGCCUUGUACUACU-UUCUCCGAUGAACAACCAGCUGUCUGCUUGAUAGCGGGUGAGUUUCGGC ..((....))................................-....((((..(((.(((.((((((.....)))))).)))..))))))). ( -18.90, z-score = -0.48, R) >droSec1.super_22 135687 91 + 1066717 AUGCUAAUGCUAUAUCUUCUAUAACCUAUGCCUUGUACUACU-UUCUCCGAUGAACAACCAGCUGUCUGCUUGAUAGCGGGUGAGUUUCGGC ..((....))................................-....((((..(((.(((.((((((.....)))))).)))..))))))). ( -18.90, z-score = -0.59, R) >droYak2.chr3R 5837413 92 - 28832112 UUGCUAAUGCUAUAUAUUCUAUACCCUAUGCCUUGUACUACUCUUCUCCAAUGAACAACCAGCUGUCUGCUUGAUAGCGGGUGAGUUUCGGU ..((....))...................(((.......(((((((......)))..(((.((((((.....)))))).)))))))...))) ( -17.60, z-score = -0.20, R) >droEre2.scaffold_4820 5925113 86 - 10470090 UUGCUAAUGCUAUAUAUUCUAUACCCUAU-----GUACUACU-UUCUCCAAUGAACAACCAGCUGUCUGCUUGAUAGCGGGUGAGUUUUGGC ..(((((.(((..........(((((...-----........-(((......)))......((((((.....)))))))))))))).))))) ( -19.80, z-score = -1.45, R) >droAna3.scaffold_13340 4502084 87 - 23697760 ----AUACUUUAUGUAUUCUACCACUUUUGACUUGUAAAUAU-UUCUUCCAUGAUCAACCAGCUGUCUGCUCGAUAGCGGGUGAGUUUUGGU ----((((.....)))).........................-.....(((.(((..(((.((((((.....)))))).)))..))).))). ( -17.60, z-score = -0.93, R) >dp4.chr2 3847686 86 + 30794189 ---AUGCUACUAUAUAUUCUAUACCUUAUG--UACUACAACU-UUCUCUUAUGAUCAACCAGCUGUCUGCUUGAUAGCGGGUGAGUUUUCGU ---.((.((.((((((..........))))--)).))))...-.......((((...(((.((((((.....)))))).)))......)))) ( -14.60, z-score = -0.33, R) >droPer1.super_7 3941910 86 - 4445127 ---AUGCUACUAUAUAUUCUAUACCUUAUG--UACUACAACU-UUCUCUUAUGAUCAACCAGCUGUCUGCUUGAUAGCGGGUGAGUUUUCGU ---.((.((.((((((..........))))--)).))))...-.......((((...(((.((((((.....)))))).)))......)))) ( -14.60, z-score = -0.33, R) >droWil1.scaffold_181089 3733831 80 + 12369635 ------AUAUUACUAUGUAUAUAUUCUAUG------AUUCAUUCUUUACUAUGAACAACCAGCUGUCUGCUCGAUAGCGGGUGAGUUUUAUC ------(((..(((..............((------.(((((........)))))))(((.((((((.....)))))).))).)))..))). ( -16.00, z-score = -1.31, R) >droVir3.scaffold_12855 8039840 74 - 10161210 ---------AU-UAUUAUAUAUCACUUAC--------UGUCUUUUAUGAACUGAACAACCAGCUGUCUGCUUGAUAGCGGGUGAGUUUUGGU ---------..-..(((((....((....--------.))....))))).(.((((.(((.((((((.....)))))).)))..)))).).. ( -16.00, z-score = -1.20, R) >droMoj3.scaffold_6540 1734598 75 - 34148556 ---------AUAAAUGAUAUCAAACUUAC--------UAUCUUUUAUGAACUGAACAACCAGCUGUCUGCUUGAUAGCGGGUGAGUUUUGGU ---------.........(((((((((((--------..((......)).........((.((((((.....))))))))))))).)))))) ( -18.10, z-score = -2.26, R) >droGri2.scaffold_15074 5619547 75 - 7742996 ---------AUACAUCAUGUAUUACUUAC--------UGUCUUUUAUGAACUAAACAACCAGCUGUCUGCUUGAUAGCGGGUGAGUUUUGGU ---------.....(((((....((....--------.))....)))))((((((..(((.((((((.....)))))).)))....)))))) ( -18.20, z-score = -1.63, R) >consensus ____UAAUGCUAUAUAUUCUAUAACCUAUG__U_GUACUACU_UUCUCCAAUGAACAACCAGCUGUCUGCUUGAUAGCGGGUGAGUUUUGGU .........................................................(((.((((((.....)))))).))).......... (-12.20 = -12.20 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:48:53 2011