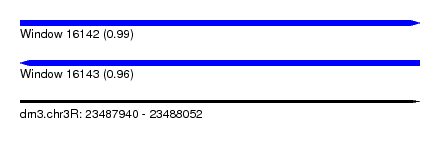
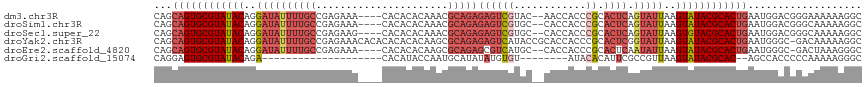
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,487,940 – 23,488,052 |
| Length | 112 |
| Max. P | 0.989925 |

| Location | 23,487,940 – 23,488,052 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.58 |
| Shannon entropy | 0.40114 |
| G+C content | 0.49429 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -27.93 |
| Energy contribution | -28.10 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
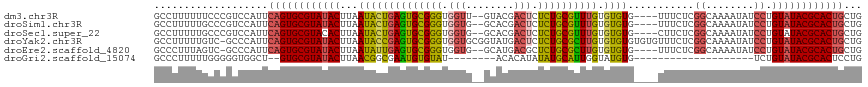
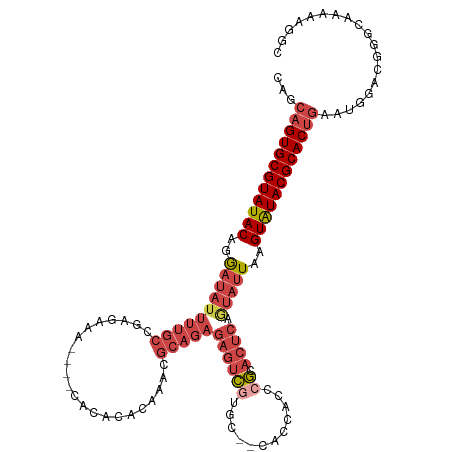
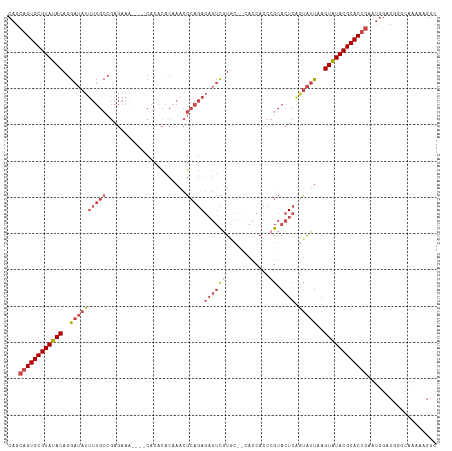
>dm3.chr3R 23487940 112 + 27905053 GCCUUUUUUCCCGUCCAUUCAGUGCGUAUACUUAAUACUGAGUGCGGGUGGUU--GUACGACUCUCUGCGUUUGUGUGUG----UUUCUCGGCAAAAUAUCCUGUAUACGCACUGCUG ...................((((((((((((...((((.((((((((..((((--....))))..)))))))))))).((----(......))).........))))))))))))... ( -32.50, z-score = -2.44, R) >droSim1.chr3R 23217775 112 + 27517382 GCCUUUUUGCCCGUCCAUUCAGUGCGUAUACUUAAUACUGAGUGCGGGUGGUG--GCACGACUCUCUGCGUUUGUGUGUG----UUUCUCGGCAAAAUAUCCUGUAUACGCACUGCUG (((...(..((((..(((((((((..((....)).)))))))))))))..).)--))..........(((...(((((((----(....(((.........))).))))))))))).. ( -34.40, z-score = -1.64, R) >droSec1.super_22 131173 112 + 1066717 GCCUUUUUGCCCGUCCAUUCAGUGCGUACACUUAAUACUGAGUGCGGGUGGUG--GCACGACUCUCUGCGUUUGUGUGUG----CUUCUCGGCAAAAUAUCCUGUAUACGCACUGCUG (((...(..((((..(((((((((...........)))))))))))))..).)--))..........(((..((((((((----(.....((........)).))))))))).))).. ( -35.20, z-score = -1.44, R) >droYak2.chr3R 5832903 117 - 28832112 GCCUUUUUGUC-GCCCAUUCAGUGCGUAUACUUAAUACCGAGUGCGGGUGGUGCGGUAUGACUCUCUGCGCUUGUGUGUGUGUGUUUCUCGGCAAAAUAUCCUGUAUACGCACUGCUG ...........-.......((((((((((((...(((((((((((((..(((........)))..))))))))).))))...(((......))).........))))))))))))... ( -39.30, z-score = -2.32, R) >droEre2.scaffold_4820 5920687 111 - 10470090 GCCCUUUAGUC-GCCCAUUCAGUGCGUAUACUUAAUAUUGAGUGCGGGUGGUG--GCAUGACGCUCUGCGCUUGUGUGUG----UUUCUCGGCAAAAUAUCCUGUAUACGCACUGCUG ...........-.......((((((((((((...(((((((((((((..((((--......)))))))))))).....((----(......))).)))))...))))))))))))... ( -35.80, z-score = -1.32, R) >droGri2.scaffold_15074 5614384 88 - 7742996 GCCCUUUUUGGGGGUGGCU--GUGCGUAUACUUAACGGCGAAUGUGUAU--------ACACAUAUAUGCAUUGGUAUGUG--------------------UCUGUAUACGCACUCCUG (((((.....)))))((..--((((((((((...(((((.(((((((((--------(....)))))))))).)).))).--------------------...)))))))))).)).. ( -35.70, z-score = -3.80, R) >consensus GCCUUUUUGCCCGUCCAUUCAGUGCGUAUACUUAAUACUGAGUGCGGGUGGUG__GCACGACUCUCUGCGUUUGUGUGUG____UUUCUCGGCAAAAUAUCCUGUAUACGCACUGCUG ...................((((((((((((...((((((((((((((.(((........))).)))))))))).))))...........((........)).))))))))))))... (-27.93 = -28.10 + 0.17)
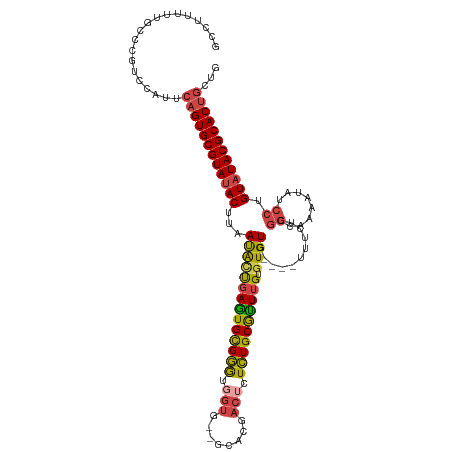
| Location | 23,487,940 – 23,488,052 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.58 |
| Shannon entropy | 0.40114 |
| G+C content | 0.49429 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -19.08 |
| Energy contribution | -21.25 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23487940 112 - 27905053 CAGCAGUGCGUAUACAGGAUAUUUUGCCGAGAAA----CACACACAAACGCAGAGAGUCGUAC--AACCACCCGCACUCAGUAUUAAGUAUACGCACUGAAUGGACGGGAAAAAAGGC ...((((((((((((..((((((((((.......----...........)))))((((((...--.......)).)))).)))))..))))))))))))................... ( -28.87, z-score = -2.53, R) >droSim1.chr3R 23217775 112 - 27517382 CAGCAGUGCGUAUACAGGAUAUUUUGCCGAGAAA----CACACACAAACGCAGAGAGUCGUGC--CACCACCCGCACUCAGUAUUAAGUAUACGCACUGAAUGGACGGGCAAAAAGGC ...((((((((((((..(((.((((((.......----...........)))))).)))((((--........))))..........))))))))))))................... ( -31.07, z-score = -2.06, R) >droSec1.super_22 131173 112 - 1066717 CAGCAGUGCGUAUACAGGAUAUUUUGCCGAGAAG----CACACACAAACGCAGAGAGUCGUGC--CACCACCCGCACUCAGUAUUAAGUGUACGCACUGAAUGGACGGGCAAAAAGGC ...((((((((((((..(((.((((((.......----...........)))))).)))((((--........))))..........))))))))))))................... ( -31.07, z-score = -1.09, R) >droYak2.chr3R 5832903 117 + 28832112 CAGCAGUGCGUAUACAGGAUAUUUUGCCGAGAAACACACACACACAAGCGCAGAGAGUCAUACCGCACCACCCGCACUCGGUAUUAAGUAUACGCACUGAAUGGGC-GACAAAAAGGC ...((((((((((((..(((.(((((((...................).)))))).)))((((((.............))))))...)))))))))))).......-........... ( -31.23, z-score = -2.24, R) >droEre2.scaffold_4820 5920687 111 + 10470090 CAGCAGUGCGUAUACAGGAUAUUUUGCCGAGAAA----CACACACAAGCGCAGAGCGUCAUGC--CACCACCCGCACUCAAUAUUAAGUAUACGCACUGAAUGGGC-GACUAAAGGGC ...((((((((((((..((((((.....(((...----.........((((...))))..(((--........))))))))))))..))))))))))))..(((..-..)))...... ( -31.00, z-score = -1.83, R) >droGri2.scaffold_15074 5614384 88 + 7742996 CAGGAGUGCGUAUACAGA--------------------CACAUACCAAUGCAUAUAUGUGU--------AUACACAUUCGCCGUUAAGUAUACGCAC--AGCCACCCCCAAAAAGGGC ..((.((((((((((.((--------------------(.(((....))).....(((((.--------...))))).....)))..))))))))))--..))...(((.....))). ( -24.80, z-score = -2.86, R) >consensus CAGCAGUGCGUAUACAGGAUAUUUUGCCGAGAAA____CACACACAAACGCAGAGAGUCGUGC__CACCACCCGCACUCAGUAUUAAGUAUACGCACUGAAUGGACGGGCAAAAAGGC ...((((((((((((..((((((((((......................)))))((((((............)).)))).)))))..))))))))))))................... (-19.08 = -21.25 + 2.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:48:50 2011