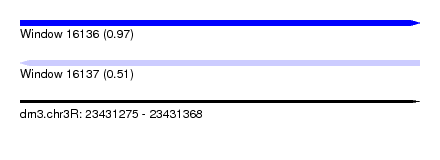
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,431,275 – 23,431,368 |
| Length | 93 |
| Max. P | 0.970494 |

| Location | 23,431,275 – 23,431,368 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 90.85 |
| Shannon entropy | 0.16010 |
| G+C content | 0.49448 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -26.54 |
| Energy contribution | -27.62 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
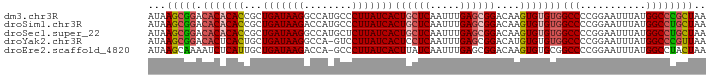
>dm3.chr3R 23431275 93 + 27905053 AUAAGCGGACACACACCGCUGAUAAGGCCAUGCCCUUAUCACUGCUCAAUUUGAGCGGACAAGUGUGUGGCCCCGGAAUUUAUGGCCCGCUAA ...(((((.(((((((...((((((((......))))))))((((((.....))))))....)))))))(((...........)))))))).. ( -39.10, z-score = -4.39, R) >droSim1.chr3R 23163482 93 + 27517382 AUAAGCGGACACACACCGCUGAUAAGACCAUGCCCUUAUCACUGCUCAAUUUGAGCGGACAAGUGUGUGGCCCCGGAAUUUAUGGCCUGCUAA ...(((((.(((((((...(((((((........)))))))((((((.....))))))....)))))))(((...........)))))))).. ( -34.20, z-score = -3.78, R) >droSec1.super_22 67830 93 + 1066717 AUAAGCGGACACACACCGCUGAUAAGGCCAUGCUCUUAUCACUGCUCAAUUUGAGCGGACAAGUGUGUGGCCCCGGAAUUUAUGGCCUGCUAA ...(((((.(((((((...(((((((((...)).)))))))((((((.....))))))....)))))))(((...........)))))))).. ( -35.50, z-score = -3.36, R) >droYak2.chr3R 5775756 92 - 28832112 AUAAGCGGACACUCACUGCUGAUAAGGCCA-GUCCUUAUCACUCCUCAAUUUGAGCGGACAUGUGUGUGGCCCCGGAAUUUAUGGCCCGUUAA ...(((((.(((.(((((.((((((((...-..))))))))((.(((.....))).)).)).))).)))(((...........)))))))).. ( -26.70, z-score = -0.94, R) >droEre2.scaffold_4820 5863291 92 - 10470090 AUAAGCAAAAUCUCAUUGCUGAUAAGACCA-GCCCUUAUCACUUAUCAAUUUGAGCGGACAAGUGUGCGGCCCCGGAAUUUAUGGCCUACUAA ...(((((.......)))))((((((....-...))))))..............(((.(....).)))((((...........))))...... ( -17.20, z-score = 0.49, R) >consensus AUAAGCGGACACACACCGCUGAUAAGGCCAUGCCCUUAUCACUGCUCAAUUUGAGCGGACAAGUGUGUGGCCCCGGAAUUUAUGGCCUGCUAA ...(((((.(((((((...(((((((........)))))))((((((.....))))))....)))))))(((...........)))))))).. (-26.54 = -27.62 + 1.08)

| Location | 23,431,275 – 23,431,368 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 90.85 |
| Shannon entropy | 0.16010 |
| G+C content | 0.49448 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -24.66 |
| Energy contribution | -25.78 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23431275 93 - 27905053 UUAGCGGGCCAUAAAUUCCGGGGCCACACACUUGUCCGCUCAAAUUGAGCAGUGAUAAGGGCAUGGCCUUAUCAGCGGUGUGUGUCCGCUUAU ..(((((((((((....(((((((((....(((((((((((.....)))).).))))))....))))))......))))))).)))))))... ( -38.00, z-score = -2.93, R) >droSim1.chr3R 23163482 93 - 27517382 UUAGCAGGCCAUAAAUUCCGGGGCCACACACUUGUCCGCUCAAAUUGAGCAGUGAUAAGGGCAUGGUCUUAUCAGCGGUGUGUGUCCGCUUAU ..(((.((((...........))))((((((....((((((.....))))..((((((((......))))))))..)).))))))..)))... ( -31.80, z-score = -1.78, R) >droSec1.super_22 67830 93 - 1066717 UUAGCAGGCCAUAAAUUCCGGGGCCACACACUUGUCCGCUCAAAUUGAGCAGUGAUAAGAGCAUGGCCUUAUCAGCGGUGUGUGUCCGCUUAU ..(((.((((...........))))((((((....((((((.....))))..(((((((.((...)))))))))..)).))))))..)))... ( -32.60, z-score = -2.12, R) >droYak2.chr3R 5775756 92 + 28832112 UUAACGGGCCAUAAAUUCCGGGGCCACACACAUGUCCGCUCAAAUUGAGGAGUGAUAAGGAC-UGGCCUUAUCAGCAGUGAGUGUCCGCUUAU ....((((........))))((((.((.(((.(((..((((........))))(((((((..-...))))))).)))))).))))))...... ( -27.00, z-score = -0.77, R) >droEre2.scaffold_4820 5863291 92 + 10470090 UUAGUAGGCCAUAAAUUCCGGGGCCGCACACUUGUCCGCUCAAAUUGAUAAGUGAUAAGGGC-UGGUCUUAUCAGCAAUGAGAUUUUGCUUAU ...((.((((...........)))))).((((((((..........))))))))....((((-.((((((((.....))))))))..)))).. ( -23.70, z-score = -0.51, R) >consensus UUAGCAGGCCAUAAAUUCCGGGGCCACACACUUGUCCGCUCAAAUUGAGCAGUGAUAAGGGCAUGGCCUUAUCAGCGGUGUGUGUCCGCUUAU ..(((((((((((......(((((((....(((((((((((.....)))).).))))))....)))))))........)))).)))))))... (-24.66 = -25.78 + 1.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:48:45 2011