| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,405,068 – 23,405,162 |
| Length | 94 |
| Max. P | 0.922221 |

| Location | 23,405,068 – 23,405,162 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 72.87 |
| Shannon entropy | 0.49504 |
| G+C content | 0.45201 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -15.14 |
| Energy contribution | -15.39 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
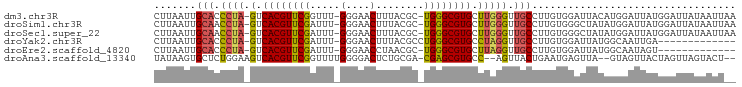
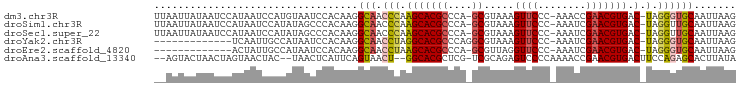
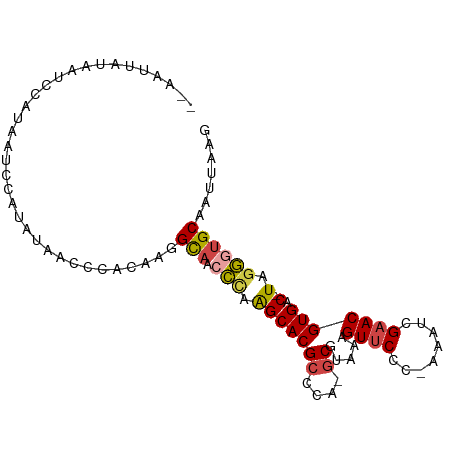
>dm3.chr3R 23405068 94 + 27905053 CUUAAUUGCACCCUA-GUCACGUUCGGUUU-GGGAACUUUACGC-UGGGCGUGCUUGGGUUGCCUUGUGGAUUACAUGGAUUAUGGAUUAUAAUUAA ..(((((((((((.(-(.((((((((((..-(....).....))-)))))))))).))).))).(..((.....))..)......)))))....... ( -26.40, z-score = -1.57, R) >droSim1.chr3R 23136351 94 + 27517382 CUUAAUUGCAACCUA-GUCACGUUCGAUUU-GGGAACUUUACGC-UGGGCGUGCUUGGGUUGCCUUGUGGGCUAUAUGGAUUAUGGAUUAUAAUUAA .......((((((((-(.((((((((....-(....).......-)))))))).))))))))).((((((.(((((.....))))).)))))).... ( -27.00, z-score = -2.14, R) >droSec1.super_22 38648 94 + 1066717 CUUAAUUGCAACCUA-GUCACGUUCGAUUU-GGGAACUUUACGC-UGGGCGUGCUUGGGUUGCCUUGUGGGCUAUAUGGAUUAUGGAUUAUAAUUAA .......((((((((-(.((((((((....-(....).......-)))))))).))))))))).((((((.(((((.....))))).)))))).... ( -27.00, z-score = -2.14, R) >droYak2.chr3R 5749026 82 - 28832112 CUUAAUUGCACCCUA-GUCACGUUCGAUUU-GGGAACUUUACGCCUGGGCGUGCCUAGGUUGCCUUGUGGAUUAUGGCAAUUGA------------- .((((((((......-.(((((..((((((-(((.....(((((....))))))))))))))...)))))......))))))))------------- ( -26.82, z-score = -1.99, R) >droEre2.scaffold_4820 5837286 81 - 10470090 CUUAAUUGCACCCUA-GUCACGUUCGAUUU-GGGAACCUAACGC-UGGGCGUGCUUAGGUUGCCUUGUGGAUUAUGGCAAUAGU------------- .......((((((((-((...((((.....-..)))).....))-)))).))))....((((((...........))))))...------------- ( -23.70, z-score = -0.92, R) >droAna3.scaffold_13340 4409907 90 - 23697760 UAUAAGUGCUCUGGAAGUCACGUUCGGUUUUGGGGACUCUGCGA-CGAGCGUGCC--AGUUACUGAAUGAGUUA--GUAGUUACUAGUUAGUACU-- ....((((((((((..(.((((((((((...(....)...))..-)))))))).)--..(((((((.....)))--))))...))))..))))))-- ( -26.40, z-score = -0.88, R) >consensus CUUAAUUGCACCCUA_GUCACGUUCGAUUU_GGGAACUUUACGC_UGGGCGUGCUUGGGUUGCCUUGUGGAUUAUAGGAAUUAUGGAUUAUAAUU__ .......(((.((((.(.((((((((.....(....)........))))))))).)))).))).................................. (-15.14 = -15.39 + 0.25)
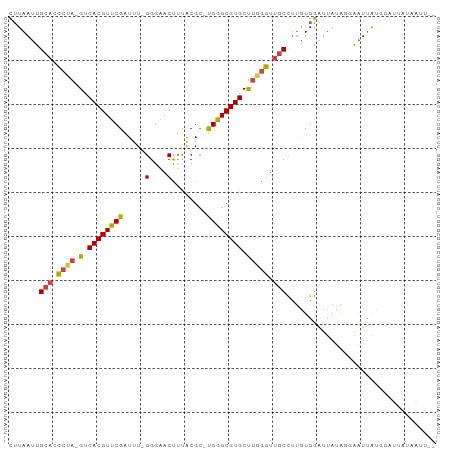
| Location | 23,405,068 – 23,405,162 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 72.87 |
| Shannon entropy | 0.49504 |
| G+C content | 0.45201 |
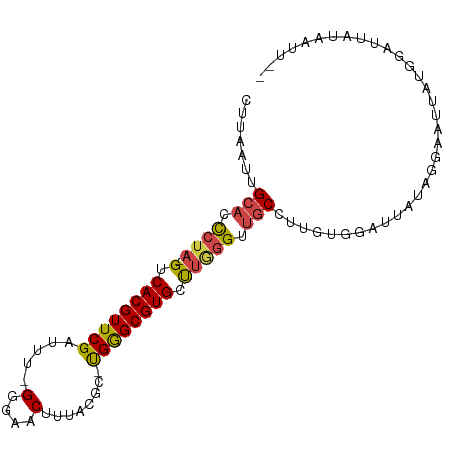
| Mean single sequence MFE | -17.60 |
| Consensus MFE | -9.77 |
| Energy contribution | -10.47 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.878236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23405068 94 - 27905053 UUAAUUAUAAUCCAUAAUCCAUGUAAUCCACAAGGCAACCCAAGCACGCCCA-GCGUAAAGUUCCC-AAACCGAACGUGAC-UAGGGUGCAAUUAAG ((((((...............(((.....)))..(((.(((.(((((((...-)).....((((..-.....))))))).)-).)))))))))))). ( -17.50, z-score = -1.77, R) >droSim1.chr3R 23136351 94 - 27517382 UUAAUUAUAAUCCAUAAUCCAUAUAGCCCACAAGGCAACCCAAGCACGCCCA-GCGUAAAGUUCCC-AAAUCGAACGUGAC-UAGGUUGCAAUUAAG ..................................((((((..(((((((...-)).....((((..-.....))))))).)-).))))))....... ( -16.20, z-score = -2.16, R) >droSec1.super_22 38648 94 - 1066717 UUAAUUAUAAUCCAUAAUCCAUAUAGCCCACAAGGCAACCCAAGCACGCCCA-GCGUAAAGUUCCC-AAAUCGAACGUGAC-UAGGUUGCAAUUAAG ..................................((((((..(((((((...-)).....((((..-.....))))))).)-).))))))....... ( -16.20, z-score = -2.16, R) >droYak2.chr3R 5749026 82 + 28832112 -------------UCAAUUGCCAUAAUCCACAAGGCAACCUAGGCACGCCCAGGCGUAAAGUUCCC-AAAUCGAACGUGAC-UAGGGUGCAAUUAAG -------------..(((((((.....((....))...(((((.(((((....)).....((((..-.....))))))).)-))))).))))))... ( -22.20, z-score = -2.17, R) >droEre2.scaffold_4820 5837286 81 + 10470090 -------------ACUAUUGCCAUAAUCCACAAGGCAACCUAAGCACGCCCA-GCGUUAGGUUCCC-AAAUCGAACGUGAC-UAGGGUGCAAUUAAG -------------....(((((...........))))).....((((.((..-..((((.((((..-.....)))).))))-..))))))....... ( -20.30, z-score = -1.73, R) >droAna3.scaffold_13340 4409907 90 + 23697760 --AGUACUAACUAGUAACUAC--UAACUCAUUCAGUAACU--GGCACGCUCG-UCGCAGAGUCCCCAAAACCGAACGUGACUUCCAGAGCACUUAUA --........(((((.(((..--..........))).)))--))...(((((-((((...................))))).....))))....... ( -13.21, z-score = 0.67, R) >consensus __AAUUAUAAUCCAUAAUCCAUAUAACCCACAAGGCAACCCAAGCACGCCCA_GCGUAAAGUUCCC_AAAUCGAACGUGAC_UAGGGUGCAAUUAAG ..................................(((.((....(((((....)).....((((........))))))).....)).)))....... ( -9.77 = -10.47 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:48:42 2011