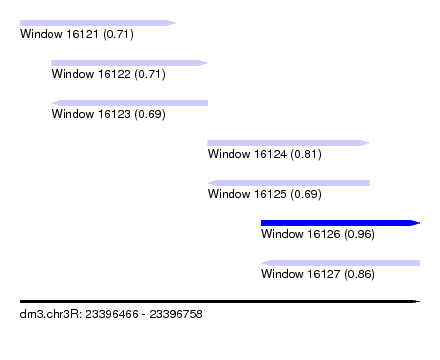
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,396,466 – 23,396,758 |
| Length | 292 |
| Max. P | 0.964779 |

| Location | 23,396,466 – 23,396,580 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 96.14 |
| Shannon entropy | 0.06838 |
| G+C content | 0.50000 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -28.54 |
| Energy contribution | -28.54 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.709561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23396466 114 + 27905053 GCUUGCUCGAUGUUUUCUGCUGGCAUUUAUAAGAUUUUCGCCGCCCAACCACUGGAAUACAGUCAUCGUUUGGUCAUCUAAUGACUAAGCGCCAGUCGGCAGCCGUCGCCCACU ...((..(((((....((((((((..........................((((.....))))...(((((((((((...)))))))))))...)))))))).)))))..)).. ( -32.70, z-score = -1.54, R) >droSim1.chr3R 23127696 114 + 27517382 GCUUGCUCGAUGUUUUCUGCUGGCAUUUAUAAGAUUUUCGCCGCCCAACCACUGGAAUACAGUCAUCGUUUGGUCAUCUAAUGACUAAGCGCCAGUCGGCAGCCAUCGCCCACU ...((..(((((....((((((((..........................((((.....))))...(((((((((((...)))))))))))...)))))))).)))))..)).. ( -33.10, z-score = -1.98, R) >droSec1.super_22 30193 114 + 1066717 GCUUGCUCGAUGUUUUCUGCUGGCAUUUAUAAGAUUUUCGCCGCCCAACCACUGGAAUACAGUCAUCGUUUGGUCAUCUAAUGACUAAGCGCCAGUCGGCAGCCAUCGCCCACU ...((..(((((....((((((((..........................((((.....))))...(((((((((((...)))))))))))...)))))))).)))))..)).. ( -33.10, z-score = -1.98, R) >droYak2.chr3R 5740150 114 - 28832112 GCUUGCCCGUUGUAUUCGGCUGGCAUUUAUAAGAUUUUCGACGCCCAACCAUUGGAAUACAGUCAUCGUUUGGUCAUCUAAUGACUAAGCGCCAGUCGGCAGCCUUCGCCCACU ....((..(((((...((((((((...............(((..(((.....)))......)))...((((((((((...)))))))))))))))))))))))....))..... ( -34.10, z-score = -2.10, R) >droEre2.scaffold_4820 5828548 114 - 10470090 GCUUGCCCGUUGUUUUCUGCUGGCAUUUAUAAGAUUUUAGCCGCCCAACCACUGGAAUACAGUCAUCGUUUGGUCAUCUAAUGACUAAGCGCCAGUAGGCAGCCUUCGCCCACU ....((..(((((...((((((((..........................((((.....))))....((((((((((...)))))))))))))))))))))))....))..... ( -32.90, z-score = -1.97, R) >consensus GCUUGCUCGAUGUUUUCUGCUGGCAUUUAUAAGAUUUUCGCCGCCCAACCACUGGAAUACAGUCAUCGUUUGGUCAUCUAAUGACUAAGCGCCAGUCGGCAGCCAUCGCCCACU (((.(((.((.....)).((((((..........................((((.....))))....((((((((((...)))))))))))))))).))))))........... (-28.54 = -28.54 + -0.00)

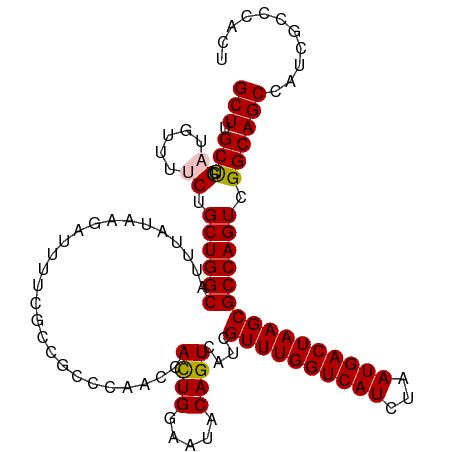
| Location | 23,396,489 – 23,396,603 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 96.49 |
| Shannon entropy | 0.06205 |
| G+C content | 0.47544 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -26.32 |
| Energy contribution | -26.96 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.713461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23396489 114 + 27905053 CAUUUAUAAGAUUUUCGCCGCCCAACCACUGGAAUACAGUCAUCGUUUGGUCAUCUAAUGACUAAGCGCCAGUCGGCAGCCGUCGCCCACUCAUCGGUGGGAAAUUAAUUUCCC .........(((....((.(((.....((((.....))))...(((((((((((...)))))))))))......))).)).))).((((((....))))))............. ( -31.40, z-score = -2.07, R) >droSim1.chr3R 23127719 114 + 27517382 CAUUUAUAAGAUUUUCGCCGCCCAACCACUGGAAUACAGUCAUCGUUUGGUCAUCUAAUGACUAAGCGCCAGUCGGCAGCCAUCGCCCACUCAUCGCUGGGAAAUUAAUUUCCC .........(((....((.(((.....((((.....))))...(((((((((((...)))))))))))......))).)).)))..............((((((....)))))) ( -27.80, z-score = -1.61, R) >droSec1.super_22 30216 114 + 1066717 CAUUUAUAAGAUUUUCGCCGCCCAACCACUGGAAUACAGUCAUCGUUUGGUCAUCUAAUGACUAAGCGCCAGUCGGCAGCCAUCGCCCACUCAUCGCUGGGAAAUUAAUUUCCC .........(((....((.(((.....((((.....))))...(((((((((((...)))))))))))......))).)).)))..............((((((....)))))) ( -27.80, z-score = -1.61, R) >droYak2.chr3R 5740173 114 - 28832112 CAUUUAUAAGAUUUUCGACGCCCAACCAUUGGAAUACAGUCAUCGUUUGGUCAUCUAAUGACUAAGCGCCAGUCGGCAGCCUUCGCCCACUCAUUGGUGGGAAAUUAAUUUCCC .........((..((.....((((.(((.((............(((((((((((...)))))))))))..(((.(((.......))).))))).))))))).....))..)).. ( -28.80, z-score = -1.54, R) >droEre2.scaffold_4820 5828571 114 - 10470090 CAUUUAUAAGAUUUUAGCCGCCCAACCACUGGAAUACAGUCAUCGUUUGGUCAUCUAAUGACUAAGCGCCAGUAGGCAGCCUUCGCCCACUCAUUGGGGGGAAAUUAAUUUCCC .........((..(((.((.(((((..((((.....))))...(((((((((((...)))))))))))..(((.(((.......))).)))..))))).))....)))..)).. ( -32.10, z-score = -1.94, R) >consensus CAUUUAUAAGAUUUUCGCCGCCCAACCACUGGAAUACAGUCAUCGUUUGGUCAUCUAAUGACUAAGCGCCAGUCGGCAGCCAUCGCCCACUCAUCGGUGGGAAAUUAAUUUCCC .........(((((..((.(((.....((((.....))))...(((((((((((...)))))))))))......))).)).....((((((....)))))))))))........ (-26.32 = -26.96 + 0.64)

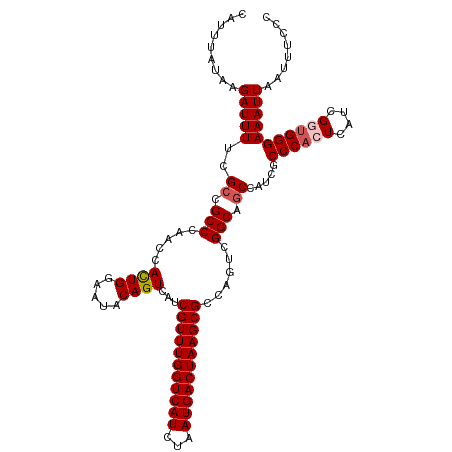
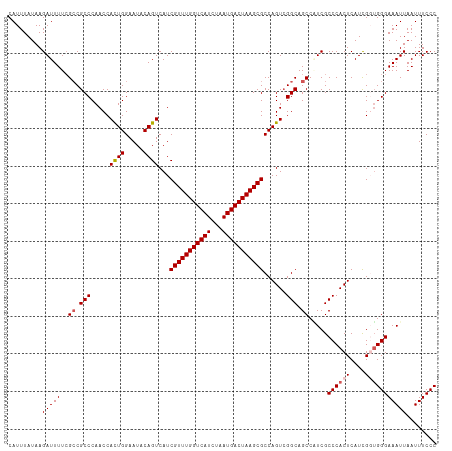
| Location | 23,396,489 – 23,396,603 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 96.49 |
| Shannon entropy | 0.06205 |
| G+C content | 0.47544 |
| Mean single sequence MFE | -34.86 |
| Consensus MFE | -31.64 |
| Energy contribution | -31.84 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.693830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23396489 114 - 27905053 GGGAAAUUAAUUUCCCACCGAUGAGUGGGCGACGGCUGCCGACUGGCGCUUAGUCAUUAGAUGACCAAACGAUGACUGUAUUCCAGUGGUUGGGCGGCGAAAAUCUUAUAAAUG ((((((....))))))...(((.(((.((((.....)))).)))(.((((..(((((...)))))....((((.((((.....)))).)))))))).)....)))......... ( -34.80, z-score = -1.40, R) >droSim1.chr3R 23127719 114 - 27517382 GGGAAAUUAAUUUCCCAGCGAUGAGUGGGCGAUGGCUGCCGACUGGCGCUUAGUCAUUAGAUGACCAAACGAUGACUGUAUUCCAGUGGUUGGGCGGCGAAAAUCUUAUAAAUG ((((((....))))))...(((.(((.((((.....)))).)))(.((((..(((((...)))))....((((.((((.....)))).)))))))).)....)))......... ( -34.80, z-score = -1.40, R) >droSec1.super_22 30216 114 - 1066717 GGGAAAUUAAUUUCCCAGCGAUGAGUGGGCGAUGGCUGCCGACUGGCGCUUAGUCAUUAGAUGACCAAACGAUGACUGUAUUCCAGUGGUUGGGCGGCGAAAAUCUUAUAAAUG ((((((....))))))...(((.(((.((((.....)))).)))(.((((..(((((...)))))....((((.((((.....)))).)))))))).)....)))......... ( -34.80, z-score = -1.40, R) >droYak2.chr3R 5740173 114 + 28832112 GGGAAAUUAAUUUCCCACCAAUGAGUGGGCGAAGGCUGCCGACUGGCGCUUAGUCAUUAGAUGACCAAACGAUGACUGUAUUCCAAUGGUUGGGCGUCGAAAAUCUUAUAAAUG ((((((....)))))).......(((.((((.....)))).)))((((((((..((((.((..((............))..)).))))..))))))))................ ( -32.60, z-score = -1.45, R) >droEre2.scaffold_4820 5828571 114 + 10470090 GGGAAAUUAAUUUCCCCCCAAUGAGUGGGCGAAGGCUGCCUACUGGCGCUUAGUCAUUAGAUGACCAAACGAUGACUGUAUUCCAGUGGUUGGGCGGCUAAAAUCUUAUAAAUG ((((((....))))))((((.....))))....(((((((((((((....((((((((............))))))))....)))))....))))))))............... ( -37.30, z-score = -2.32, R) >consensus GGGAAAUUAAUUUCCCACCGAUGAGUGGGCGAAGGCUGCCGACUGGCGCUUAGUCAUUAGAUGACCAAACGAUGACUGUAUUCCAGUGGUUGGGCGGCGAAAAUCUUAUAAAUG ((((((....)))))).......(((.((((.....)))).)))(.((((..(((((...)))))....((((.((((.....)))).)))))))).)................ (-31.64 = -31.84 + 0.20)

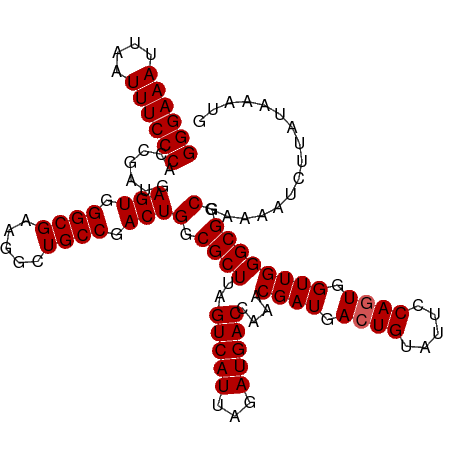
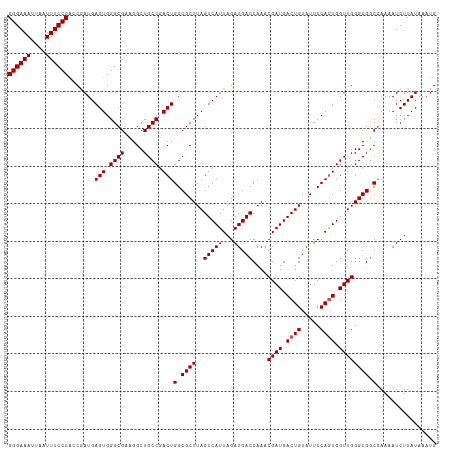
| Location | 23,396,603 – 23,396,721 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.67 |
| Shannon entropy | 0.38564 |
| G+C content | 0.41943 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -14.08 |
| Energy contribution | -15.48 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.808992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23396603 118 + 27905053 ACGAUAAGCCGCCUCUUGCAAAUUGAAGAUGGGCAAGCU--ACGACUGCCCAAUCGAUAUUACUAUCGAAAUCAUCGAUGCUUCUUUAUCGAUAAACUAAAAACACUUGAUUCCUUAGGA .((((((((.((.....))..(((((.((((((((....--.....)))))..((((((....)))))).))).))))))))....)))))..............(((((....))))). ( -25.60, z-score = -1.99, R) >droSim1.chr3R 23127833 109 + 27517382 ACCCAAAGCCACCUCUUGCAAAGUGAAGAUGGGCAAGCU--ACGACUGCCCAAUCGAUAUUUCUAUCAAAAUUAUCGACGCUUCAUUAUCGAUAAG---------CUUGAUUCCUUUAGA .......((........))((((.(((((((((((....--.....))))).)))..........((((..(((((((..........))))))).---------.)))))))))))... ( -22.10, z-score = -1.22, R) >droSec1.super_22 30330 109 + 1066717 ACCCAAAGCCGCCUCUUGCAAAUUGAAGAUGGGCAAGCU--ACGAUUGCCCAAUCGAUAUUCCUAUCGAAAUUAUCGACACUUCUUUAUCGAUAAG---------CUUGAUUCCUUAAGA .............((..((..........(((((((...--....))))))).((((((....))))))..(((((((..........))))))))---------)..)).......... ( -23.80, z-score = -1.80, R) >droYak2.chr3R 5740287 119 - 28832112 ACCCAACGCCACC-CGCCCAAAAUGGAAGUGGCAUAGCUAGAUGACUGCCCAAUCGAUAUUGCUAUCGAACUUAUCGGUGCUUCUUUAUCGAUAAACAUAAAAUACUUGAUUCUUUUAGG .......(((((.-..((......))..)))))....(((((.((........((((((....))))))..(((((((((......)))))))))................)).))))). ( -26.30, z-score = -1.88, R) >droEre2.scaffold_4820 5828685 117 - 10470090 ACCCAAAACCACC-CUCCCCAAUGGGAGGUGGCAAGGCU--ACGACUGCACAAUCGAUAUUCCUAUCGAACUUAUCGGUGCUUCUUUAUCGAUGAACCUAACAUAAUUCAUUCCUUUGGA ..(((((.(((((-.((((....)))))))))..(((..--............((((((....))))))..(((((((((......))))))))).)))...............))))). ( -33.10, z-score = -3.34, R) >consensus ACCCAAAGCCACCUCUUGCAAAUUGAAGAUGGGCAAGCU__ACGACUGCCCAAUCGAUAUUCCUAUCGAAAUUAUCGAUGCUUCUUUAUCGAUAAAC__AA_A_ACUUGAUUCCUUUAGA .......................(((((.((((((..(.....)..)))))).((((((....))))))..(((((((((......)))))))))..................))))).. (-14.08 = -15.48 + 1.40)

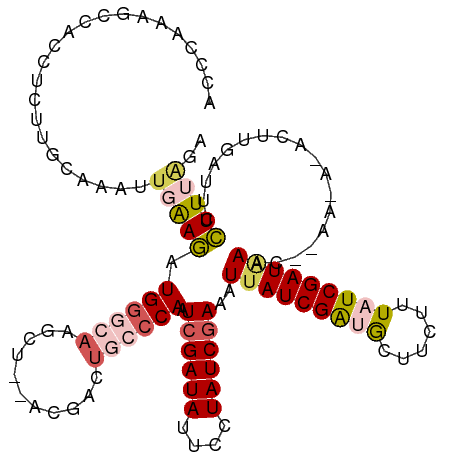
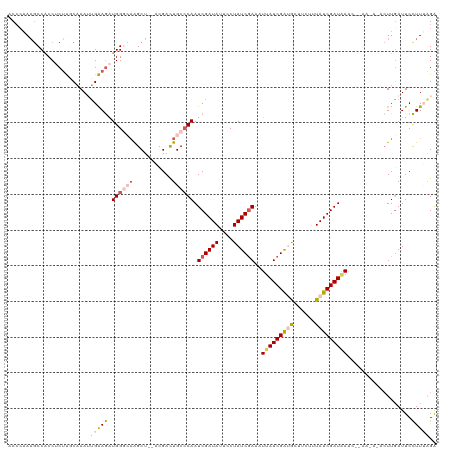
| Location | 23,396,603 – 23,396,721 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.67 |
| Shannon entropy | 0.38564 |
| G+C content | 0.41943 |
| Mean single sequence MFE | -32.39 |
| Consensus MFE | -19.56 |
| Energy contribution | -20.88 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.694712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23396603 118 - 27905053 UCCUAAGGAAUCAAGUGUUUUUAGUUUAUCGAUAAAGAAGCAUCGAUGAUUUCGAUAGUAAUAUCGAUUGGGCAGUCGU--AGCUUGCCCAUCUUCAAUUUGCAAGAGGCGGCUUAUCGU .((((((((((((.((((((((.............))))))))...)))))))((((....))))..))))).((((((--..(((((.............)))))..))))))...... ( -31.54, z-score = -1.72, R) >droSim1.chr3R 23127833 109 - 27517382 UCUAAAGGAAUCAAG---------CUUAUCGAUAAUGAAGCGUCGAUAAUUUUGAUAGAAAUAUCGAUUGGGCAGUCGU--AGCUUGCCCAUCUUCACUUUGCAAGAGGUGGCUUUGGGU (((((((..((((((---------.((((((((........)))))))).)))))).........(((.(((((((...--.)).)))))))).(((((((....)))))))))))))). ( -33.60, z-score = -2.44, R) >droSec1.super_22 30330 109 - 1066717 UCUUAAGGAAUCAAG---------CUUAUCGAUAAAGAAGUGUCGAUAAUUUCGAUAGGAAUAUCGAUUGGGCAAUCGU--AGCUUGCCCAUCUUCAAUUUGCAAGAGGCGGCUUUGGGU .(((((((......(---------((((((((((......((((((.....))))))....)))))).)))))..((((--..(((((.............)))))..))))))))))). ( -30.12, z-score = -1.40, R) >droYak2.chr3R 5740287 119 + 28832112 CCUAAAAGAAUCAAGUAUUUUAUGUUUAUCGAUAAAGAAGCACCGAUAAGUUCGAUAGCAAUAUCGAUUGGGCAGUCAUCUAGCUAUGCCACUUCCAUUUUGGGCG-GGUGGCGUUGGGU (((((...................(((((((............))))))).((((((....))))))))))).....(((((((...(((((((((......)).)-))))))))))))) ( -30.70, z-score = -0.96, R) >droEre2.scaffold_4820 5828685 117 + 10470090 UCCAAAGGAAUGAAUUAUGUUAGGUUCAUCGAUAAAGAAGCACCGAUAAGUUCGAUAGGAAUAUCGAUUGUGCAGUCGU--AGCCUUGCCACCUCCCAUUGGGGAG-GGUGGUUUUGGGU ((((((((.(((((((......)))))))..........(((((((((..(((....))).)))))...))))......--..))))((((((((((....)))).-))))))..)))). ( -36.00, z-score = -1.77, R) >consensus UCUAAAGGAAUCAAGU_U_UU__GUUUAUCGAUAAAGAAGCAUCGAUAAUUUCGAUAGGAAUAUCGAUUGGGCAGUCGU__AGCUUGCCCAUCUUCAAUUUGCAAGAGGUGGCUUUGGGU (((((((..................((((((((........))))))))..((((((....)))))).(((((((.(.....).))))))).....................))))))). (-19.56 = -20.88 + 1.32)

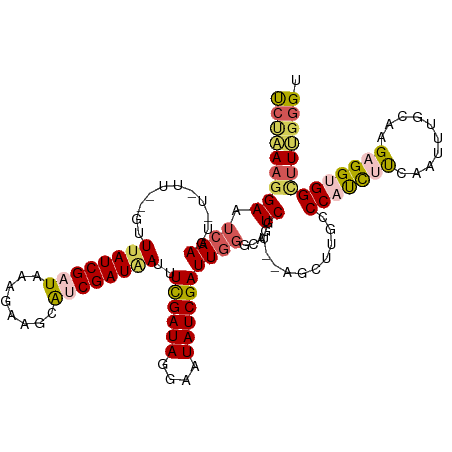
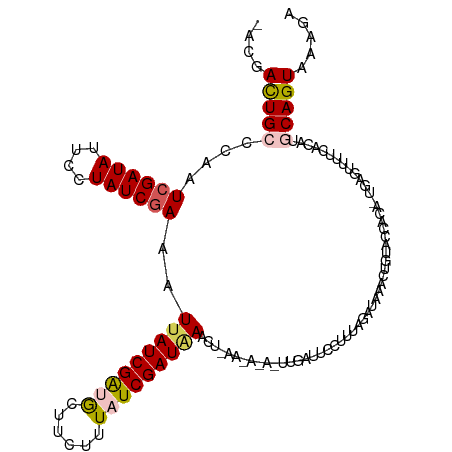
| Location | 23,396,642 – 23,396,758 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.30 |
| Shannon entropy | 0.43285 |
| G+C content | 0.33588 |
| Mean single sequence MFE | -19.14 |
| Consensus MFE | -14.10 |
| Energy contribution | -14.62 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23396642 116 + 27905053 -ACGACUGCCCAAUCGAUAUUACUAUCGAAAUCAUCGAUGCUUCUUUAUCGAUAAACUAAAAACACUUGAUUCCUUAGGAUAAACAGUACCACA-UGAGUUUUUACAUGCAGUAAAGA -...(((((...(((((((....((((((.....))))))......)))))))....(((((((.(((((....))))).....((........-)).)))))))...)))))..... ( -22.60, z-score = -2.60, R) >droSim1.chr3R 23127872 107 + 27517382 -ACGACUGCCCAAUCGAUAUUUCUAUCAAAAUUAUCGACGCUUCAUUAUCGAUAAGCU---------UGAUUCCUUUAGAUAAACUGUACCACA-UGAGUUUUCACAUGCAGUAAAGA -...(((((...............(((((..(((((((..........)))))))..)---------)))).......((.(((((........-..)))))))....)))))..... ( -17.30, z-score = -1.00, R) >droSec1.super_22 30369 98 + 1066717 -ACGAUUGCCCAAUCGAUAUUCCUAUCGAAAUUAUCGACACUUCUUUAUCGAUAAGCU---------UGAUUCCUUAAGAUAAACUG----------AGUUUUCACAUGCAGUAAAGA -...(((((....((((((....)))))).......((.((((.((((((..((((..---------......)))).))))))..)----------)))..))....)))))..... ( -16.00, z-score = -0.98, R) >droYak2.chr3R 5740326 117 - 28832112 GAUGACUGCCCAAUCGAUAUUGCUAUCGAACUUAUCGGUGCUUCUUUAUCGAUAAACAUAAAAUACUUGAUUCUUUUAGGUAAACUGUACUUUUCUUUA-GUUUUUAAACAGUAAAUA ....((((....(((((((..((.(((((.....))))))).....)))))))..........(((((((.....)))))))(((((..........))-)))......))))..... ( -20.50, z-score = -1.80, R) >droEre2.scaffold_4820 5828723 117 - 10470090 -ACGACUGCACAAUCGAUAUUCCUAUCGAACUUAUCGGUGCUUCUUUAUCGAUGAACCUAACAUAAUUCAUUCCUUUGGAUAAACUGUACUUUUCUUUACGUUAACAAACAGUAAAUA -...((((.....((((((....))))))..(((((((((......)))))))))...((((...(((((......))))).....(((........))))))).....))))..... ( -19.30, z-score = -2.13, R) >consensus _ACGACUGCCCAAUCGAUAUUCCUAUCGAAAUUAUCGAUGCUUCUUUAUCGAUAAACU_AA_A_A_UUGAUUCCUUUAGAUAAACUGUACCACA_UGAGUUUUCACAUGCAGUAAAGA ....(((((....((((((....))))))..(((((((((......))))))))).....................................................)))))..... (-14.10 = -14.62 + 0.52)

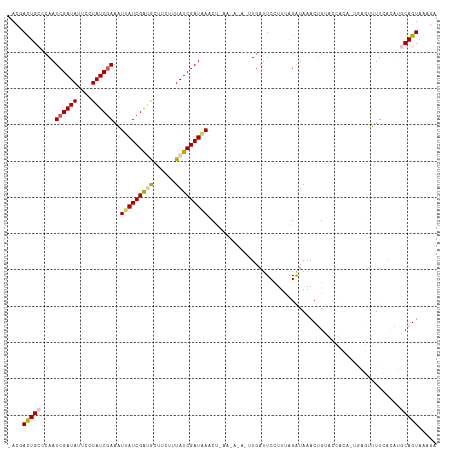
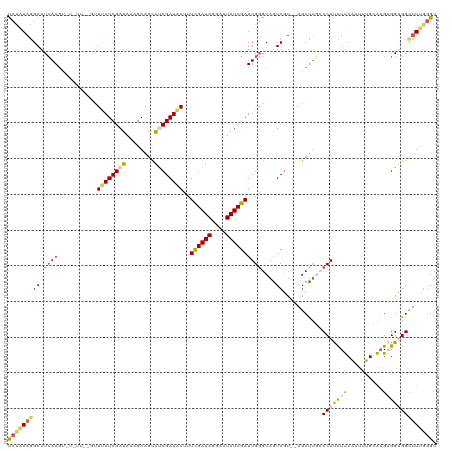
| Location | 23,396,642 – 23,396,758 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.30 |
| Shannon entropy | 0.43285 |
| G+C content | 0.33588 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -15.68 |
| Energy contribution | -15.72 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23396642 116 - 27905053 UCUUUACUGCAUGUAAAAACUCA-UGUGGUACUGUUUAUCCUAAGGAAUCAAGUGUUUUUAGUUUAUCGAUAAAGAAGCAUCGAUGAUUUCGAUAGUAAUAUCGAUUGGGCAGUCGU- ....(((..((((........))-))..)))(((((((..(((((((........))))))).((((((((........))))))))..((((((....)))))).)))))))....- ( -30.30, z-score = -2.65, R) >droSim1.chr3R 23127872 107 - 27517382 UCUUUACUGCAUGUGAAAACUCA-UGUGGUACAGUUUAUCUAAAGGAAUCA---------AGCUUAUCGAUAAUGAAGCGUCGAUAAUUUUGAUAGAAAUAUCGAUUGGGCAGUCGU- .((.(((..((((........))-))..))).)).............((((---------((.((((((((........)))))))).))))))........(((((....))))).- ( -25.80, z-score = -1.63, R) >droSec1.super_22 30369 98 - 1066717 UCUUUACUGCAUGUGAAAACU----------CAGUUUAUCUUAAGGAAUCA---------AGCUUAUCGAUAAAGAAGUGUCGAUAAUUUCGAUAGGAAUAUCGAUUGGGCAAUCGU- (((((((((...((....)).----------)))).......)))))....---------.(((((((((((......((((((.....))))))....)))))).)))))......- ( -20.61, z-score = -0.85, R) >droYak2.chr3R 5740326 117 + 28832112 UAUUUACUGUUUAAAAAC-UAAAGAAAAGUACAGUUUACCUAAAAGAAUCAAGUAUUUUAUGUUUAUCGAUAAAGAAGCACCGAUAAGUUCGAUAGCAAUAUCGAUUGGGCAGUCAUC .....(((((((((....-......(((((((.((((........))))...)))))))...(((((((............))))))).((((((....))))))))))))))).... ( -20.80, z-score = -1.61, R) >droEre2.scaffold_4820 5828723 117 + 10470090 UAUUUACUGUUUGUUAACGUAAAGAAAAGUACAGUUUAUCCAAAGGAAUGAAUUAUGUUAGGUUCAUCGAUAAAGAAGCACCGAUAAGUUCGAUAGGAAUAUCGAUUGUGCAGUCGU- ..(((((.(((....))))))))((...((((((((..(((...)))..(((((.((((.(((.(.((......)).).))))))))))))((((....))))))))))))..))..- ( -22.00, z-score = -0.72, R) >consensus UCUUUACUGCAUGUAAAAACUAA_AAAAGUACAGUUUAUCUAAAGGAAUCAA_U_U_UU_AGUUUAUCGAUAAAGAAGCAUCGAUAAUUUCGAUAGGAAUAUCGAUUGGGCAGUCGU_ .....(((((.....................................................((((((((........))))))))..((((((....))))))....))))).... (-15.68 = -15.72 + 0.04)

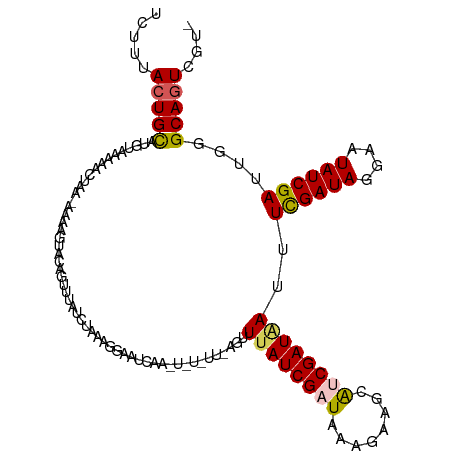
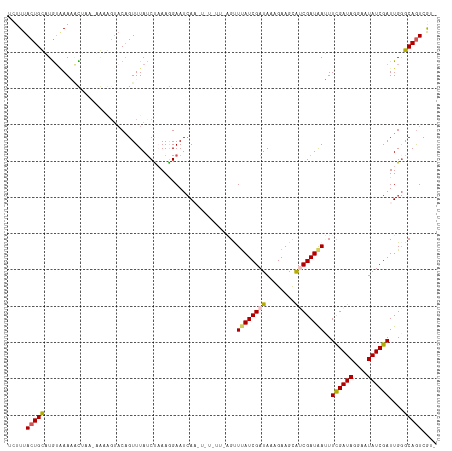
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:48:36 2011