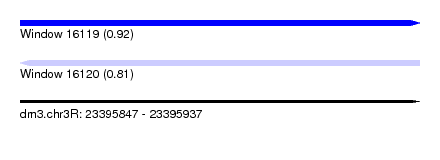
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,395,847 – 23,395,937 |
| Length | 90 |
| Max. P | 0.916095 |

| Location | 23,395,847 – 23,395,937 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 87.54 |
| Shannon entropy | 0.21581 |
| G+C content | 0.49118 |
| Mean single sequence MFE | -21.89 |
| Consensus MFE | -20.28 |
| Energy contribution | -19.80 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
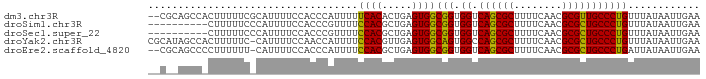
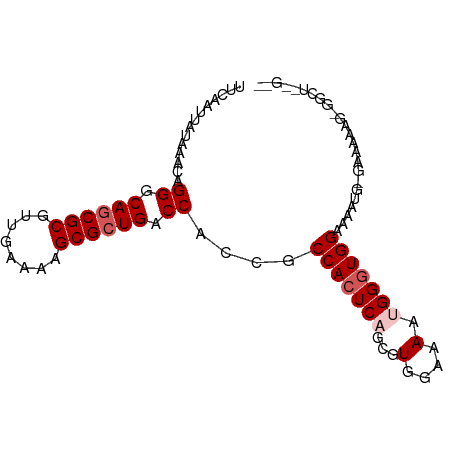
>dm3.chr3R 23395847 90 + 27905053 --CGCAGCCACUUUUUCGCAUUUUCCACCCAUUUUUCACACUGAGUGGCGGUGGUCAGCGCUUUUCAACGCGUUGCCCUGUUUAUAAUUGAA --....(((((((.............................)))))))((.((.((((((........))))))))))............. ( -21.25, z-score = -0.94, R) >droSim1.chr3R 23127098 82 + 27517382 ----------CUUUUUCCCAUUUUCCACCCGUUUUCCACGCUGAGUGGCGGUGGUCAGCGCUUUUCAACGCGCUGCCCUGUUUAUAAUUGAA ----------..............(((((......((((.....)))).))))).((((((........))))))................. ( -20.80, z-score = -1.56, R) >droSec1.super_22 29585 82 + 1066717 ----------CUUUUUCCCAUUUUCCACCCGUUUUCCACGCUGAGUGGCGGUGGUCAGCGCUUUUCAACGCGCUGCCCUGUUUAUAAUUGAA ----------..............(((((......((((.....)))).))))).((((((........))))))................. ( -20.80, z-score = -1.56, R) >droYak2.chr3R 5738809 91 - 28832112 CGCAUAGCCACUUUUUC-CAUUUUCCAACCAUUUUCCACGUUGAGUGGCAGUGGCCAGCGCUUUUCAACGCGCUGCCCUGUUUAUAAUUGAA ......(((((((....-........................)))))))((.(((.(((((........))))))))))............. ( -23.69, z-score = -2.27, R) >droEre2.scaffold_4820 5827956 89 - 10470090 --CGCAGCCCCUUUUUU-CAUUUUCCACCCAUUUUCCACGCUGAGUGGCGGUGGUCAGCGCUUUUCAACGCGCUGCCCUGAUUAUAAUUGAA --.(((((.........-......(((((......((((.....)))).)))))...(((........))))))))................ ( -22.90, z-score = -1.38, R) >consensus __C__AGCC_CUUUUUC_CAUUUUCCACCCAUUUUCCACGCUGAGUGGCGGUGGUCAGCGCUUUUCAACGCGCUGCCCUGUUUAUAAUUGAA ...................................((((.....))))(((.((.((((((........)))))))))))............ (-20.28 = -19.80 + -0.48)
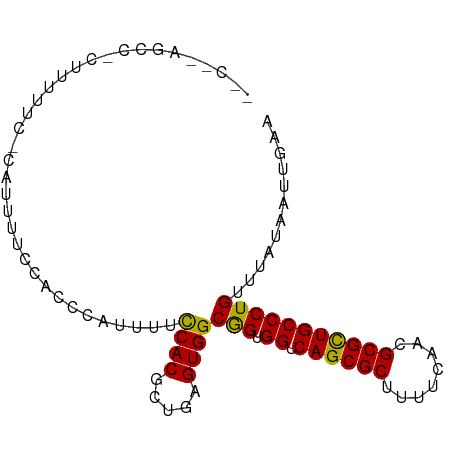
| Location | 23,395,847 – 23,395,937 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 87.54 |
| Shannon entropy | 0.21581 |
| G+C content | 0.49118 |
| Mean single sequence MFE | -25.99 |
| Consensus MFE | -21.40 |
| Energy contribution | -22.20 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.808490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23395847 90 - 27905053 UUCAAUUAUAAACAGGGCAACGCGUUGAAAAGCGCUGACCACCGCCACUCAGUGUGAAAAAUGGGUGGAAAAUGCGAAAAAGUGGCUGCG-- ................(((..(((((....)))))...((((..(((((((...(....).))))))).............)))).))).-- ( -23.06, z-score = -0.49, R) >droSim1.chr3R 23127098 82 - 27517382 UUCAAUUAUAAACAGGGCAGCGCGUUGAAAAGCGCUGACCACCGCCACUCAGCGUGGAAAACGGGUGGAAAAUGGGAAAAAG---------- (((..............((((((........)))))).(((((.((((.....))))......))))).......)))....---------- ( -24.30, z-score = -1.90, R) >droSec1.super_22 29585 82 - 1066717 UUCAAUUAUAAACAGGGCAGCGCGUUGAAAAGCGCUGACCACCGCCACUCAGCGUGGAAAACGGGUGGAAAAUGGGAAAAAG---------- (((..............((((((........)))))).(((((.((((.....))))......))))).......)))....---------- ( -24.30, z-score = -1.90, R) >droYak2.chr3R 5738809 91 + 28832112 UUCAAUUAUAAACAGGGCAGCGCGUUGAAAAGCGCUGGCCACUGCCACUCAACGUGGAAAAUGGUUGGAAAAUG-GAAAAAGUGGCUAUGCG ................(((..(((((....)))))((((((((.((((.....))))....(.(((....))).-)....))))))))))). ( -28.50, z-score = -2.18, R) >droEre2.scaffold_4820 5827956 89 + 10470090 UUCAAUUAUAAUCAGGGCAGCGCGUUGAAAAGCGCUGACCACCGCCACUCAGCGUGGAAAAUGGGUGGAAAAUG-AAAAAAGGGGCUGCG-- ................((((((((((....)))))...(((((.((((.....))))......)))))......-.........))))).-- ( -29.80, z-score = -2.32, R) >consensus UUCAAUUAUAAACAGGGCAGCGCGUUGAAAAGCGCUGACCACCGCCACUCAGCGUGGAAAAUGGGUGGAAAAUG_GAAAAAG_GGCU__G__ ..............((.((((((........)))))).))....(((((((...(....).)))))))........................ (-21.40 = -22.20 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:48:31 2011