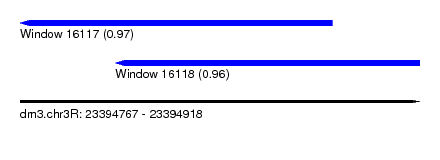
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,394,767 – 23,394,918 |
| Length | 151 |
| Max. P | 0.968832 |

| Location | 23,394,767 – 23,394,885 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 92.20 |
| Shannon entropy | 0.13502 |
| G+C content | 0.54576 |
| Mean single sequence MFE | -48.24 |
| Consensus MFE | -42.96 |
| Energy contribution | -43.00 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
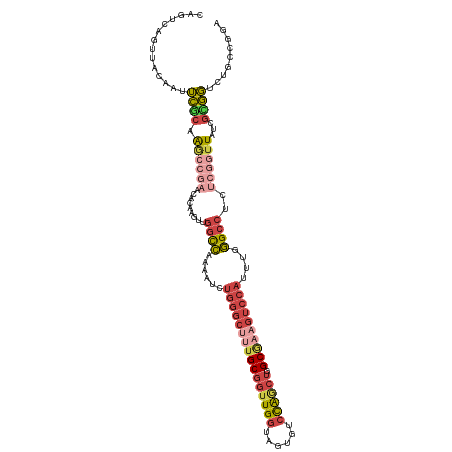
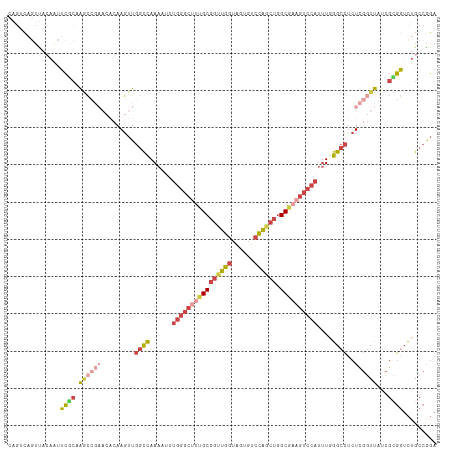
>dm3.chr3R 23394767 118 - 27905053 UGGCCAAAAGCUGGGCUUUGCGGUUGGUGGUGUCCAACUGGCGAAGUCCAUUUGGGCCUCUCGGUUAACGUGGACUGCCGGAGAAAGUUCAGCUGGCGGGAACGGAAUUCAGGACUCA .((((...((.((((((((((((((((......)))))).)))))))))).)).))))((..((....(((...(((((((.((....))..)))))))..)))....))..)).... ( -48.10, z-score = -1.90, R) >droSim1.chr3R 23126033 118 - 27517382 UGGCCAAAAUCUGGGCUUUGCGGUUGGUAGUGUCCAGCUGGCGAAGUCCAUUUGGGCCUCUCGGUUAACGCGGUCUGCCAGAGAAAGUUCAGCUGGCGGGAAUGGAAUUCAGGAUUCA .((((...(..((((((((((((((((......)))))).))))))))))..).))))((..((....((...((((((((.((....))..))))))))..))....))..)).... ( -45.10, z-score = -1.58, R) >droSec1.super_22 28518 118 - 1066717 UGGCCAAAAUCUGGGCUUUGCGGUUGGUAGUGUCCAGCUGGCGAAGUCCAUUUGGGCCUCUCGGUUAUCGCGGUCUGCCAGAGAAAGUUCAGCUGGCGGGAAUGGAAUUCAGGAUUCA .((((...(..((((((((((((((((......)))))).))))))))))..).))))(((..(((.(((...((((((((.((....))..))))))))..))))))..)))..... ( -45.40, z-score = -1.62, R) >droYak2.chr3R 5737709 118 + 28832112 UGGCCAAAUUCUGGGCUCUGCGGUUGGCAUUGUCCAGCUGGCAAUGUCCAUUUGGGCCUCUCGGUUUUCGUGGUCAGCCGGAGAAAGUUCAGCUGGCGGGAACGGAAUUCAGGAUUCA .((((......(((((..(((((((((......)))))).)))..)))))....))))((..(..((((((.((((((.(((.....))).))))))....))))))..)..)).... ( -42.90, z-score = -0.69, R) >droEre2.scaffold_4820 5826877 118 + 10470090 UGGCCAAAUUCUGGGCUUUGCGGUUGGUAGUGUCCAGCUGGCGAAGUCCAUUUUGGCCUCUUGAUUUUCGUGGUCUGCCGGAGAAAGUUCAGCUGGCGGGAACGGAAUUCAGGAUUCA .(((((((...((((((((((((((((......)))))).)))))))))).)))))))((((((.((((((..((((((((.((....))..)))))))).)))))).)))))).... ( -59.70, z-score = -6.01, R) >consensus UGGCCAAAAUCUGGGCUUUGCGGUUGGUAGUGUCCAGCUGGCGAAGUCCAUUUGGGCCUCUCGGUUAUCGUGGUCUGCCGGAGAAAGUUCAGCUGGCGGGAACGGAAUUCAGGAUUCA .((((......((((((((((((((((......)))))).))))))))))....))))(((.((....(((...(((((((.((....))..)))))))..)))....)).))).... (-42.96 = -43.00 + 0.04)

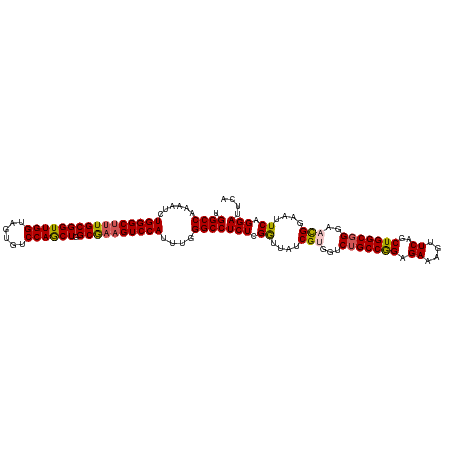
| Location | 23,394,803 – 23,394,918 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 69.83 |
| Shannon entropy | 0.58118 |
| G+C content | 0.53633 |
| Mean single sequence MFE | -38.87 |
| Consensus MFE | -24.03 |
| Energy contribution | -26.42 |
| Covariance contribution | 2.39 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23394803 115 - 27905053 CAGUCAGUUACAACUCGCAAACCGAACACAAGUUGGCCAAAAGCUGGGCUUUGCGGUUGGUGGUGUCCAACUGGCGAAGUCCAUUUGGGCCUCUCGGUUAACGUGGACUGCCGGA ((((((((....)))(((.((((((((....)).((((...((.((((((((((((((((......)))))).)))))))))).)).))))..))))))...))))))))..... ( -43.80, z-score = -1.63, R) >dp4.chr2 9041767 96 - 30794189 --GGGAAUUAAAAUGGAAUGUGU-----AAAUUUAAUUAAACGCCGAG----GCGUCUGCUGCCAUUUGCC--GCAAGGAC----UGAGCGGCU--GUUGCUGCCUUUUGCCGCA --((.((.......((...((.(-----((......))).)).))(((----(((.(.(((((((....((--....))..----)).))))).--...).)))))))).))... ( -24.20, z-score = 1.12, R) >droSim1.chr3R 23126069 114 - 27517382 CAGUCAGUUACAAUUCGC-AGCCGAACACAAGUUGGCCAAAAUCUGGGCUUUGCGGUUGGUAGUGUCCAGCUGGCGAAGUCCAUUUGGGCCUCUCGGUUAACGCGGUCUGCCAGA ................((-(((((...((.....((((...(..((((((((((((((((......)))))).))))))))))..).)))).....)).....))).)))).... ( -42.80, z-score = -2.04, R) >droSec1.super_22 28554 114 - 1066717 CAGUCAGUUACAAUUCGC-AGCCGAACACAAGUUGGCCAAAAUCUGGGCUUUGCGGUUGGUAGUGUCCAGCUGGCGAAGUCCAUUUGGGCCUCUCGGUUAUCGCGGUCUGCCAGA ................((-(((((...((.....((((...(..((((((((((((((((......)))))).))))))))))..).)))).....)).....))).)))).... ( -42.80, z-score = -2.16, R) >droYak2.chr3R 5737745 107 + 28832112 --------CAGCAGUCACAAGCCGAACACAAGUUGGCCAAAUUCUGGGCUCUGCGGUUGGCAUUGUCCAGCUGGCAAUGUCCAUUUGGGCCUCUCGGUUUUCGUGGUCAGCCGGA --------............(((.(((....))))))......((((((..(((.....)))..))))))(((((..((.((((..(((((....)))))..)))).))))))). ( -36.10, z-score = -0.24, R) >droEre2.scaffold_4820 5826913 109 + 10470090 CAGUCAGUUACAGCUCGCAAGC------GAAGUUGGCCAAAUUCUGGGCUUUGCGGUUGGUAGUGUCCAGCUGGCGAAGUCCAUUUUGGCCUCUUGAUUUUCGUGGUCUGCCGGA ..(.(((((((((((((....)------).))))(((((((...((((((((((((((((......)))))).)))))))))).)))))))...........)))).))).)... ( -43.50, z-score = -2.74, R) >consensus CAGUCAGUUACAAUUCGCAAGCCGAACACAAGUUGGCCAAAAUCUGGGCUUUGCGGUUGGUAGUGUCCAGCUGGCGAAGUCCAUUUGGGCCUCUCGGUUAUCGCGGUCUGCCGGA ..............((((.((((((.........((((......((((((((((((((((......)))))).))))))))))....))))..))))))...))))......... (-24.03 = -26.42 + 2.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:48:30 2011