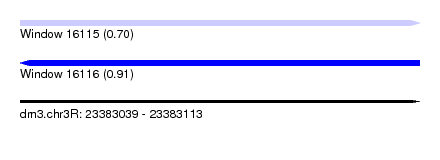
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,383,039 – 23,383,113 |
| Length | 74 |
| Max. P | 0.905218 |

| Location | 23,383,039 – 23,383,113 |
|---|---|
| Length | 74 |
| Sequences | 7 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 70.32 |
| Shannon entropy | 0.58815 |
| G+C content | 0.40381 |
| Mean single sequence MFE | -18.09 |
| Consensus MFE | -7.54 |
| Energy contribution | -7.51 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.695335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
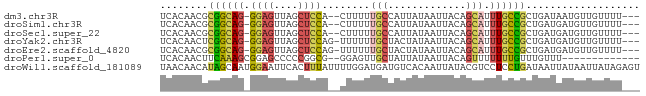
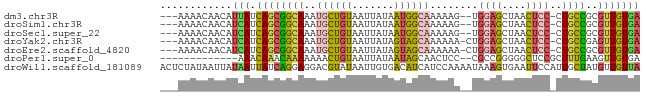
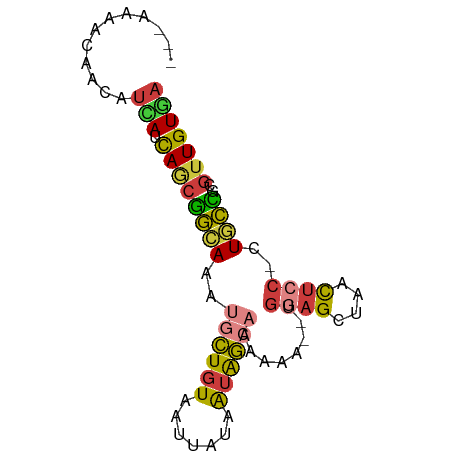
>dm3.chr3R 23383039 74 + 27905053 UCACAACGCGGCAG-GGAGUUAGCUCCA--CUUUUUGCCAUUAUAAUUACAGCAUUUGCCGCUGAUAAUGUUGUUUU--- ..(((((..(((((-(((((.......)--)))))))))......((((((((.......)))).)))))))))...--- ( -22.00, z-score = -2.80, R) >droSim1.chr3R 23114249 74 + 27517382 UCACAACGCGGCAG-GGAGUUAGCUCCA--CUUUUUGCCAUUAUAAUUACAGCAUUUGCCGCUGAUGAUGUUGUUUU--- ..(((((..(((((-(((((.......)--)))))))))......((((((((.......)))).)))))))))...--- ( -22.10, z-score = -2.49, R) >droSec1.super_22 16858 74 + 1066717 UCACAACGCGGCAG-GGAGUUAGCUCCA--CUUUUUGCCAUUAUAAUUACAGCAUUUGCCGCUGAUGAUGUUGUUUU--- ..(((((..(((((-(((((.......)--)))))))))......((((((((.......)))).)))))))))...--- ( -22.10, z-score = -2.49, R) >droYak2.chr3R 5725795 75 - 28832112 UCACAACUCGGCAG-GGAGUUAGCUCCAG-UUUUUUGCUACUAUAAUUACAGCAUUUGCCGCUGAUGAUGUUGUUUU--- ..((((((((((((-((((....))))..-....)))))..........((((.......))))..)).)))))...--- ( -18.10, z-score = -1.04, R) >droEre2.scaffold_4820 5813936 75 - 10470090 UCACAACGCGGCAG-GGAGUUAGCUCCAG-UUUUUUGCUACUAUAAUUACAGCAUUUGCCGCUGAUGAUGUUGUUUU--- ..((((((((((((-((((....))))..-.....((((...........)))).))))))).......)))))...--- ( -21.51, z-score = -1.93, R) >droPer1.super_0 2842567 65 + 11822988 UCACAACUUCAAAGCGGAGCCCCCGGCG--GGAGUUGCUAUUAUAAUUACAGUUUUUUUGUUUGUUU------------- ...(((((((...((((.....)).)).--)))))))...........((((.....))))......------------- ( -11.90, z-score = 0.07, R) >droWil1.scaffold_181089 3621508 80 + 12369635 UAACAACAUAGCAAUGGAAUUCACUUUAUUUUGGAUGAUGUCACAAUUAUACGUCCUCCUGAUAAUUAUAAUUAUAGAGU ..........((..((.((((....(((((..(((.(((((.........))))).))).)))))....)))).))..)) ( -8.90, z-score = 0.67, R) >consensus UCACAACGCGGCAG_GGAGUUAGCUCCA__CUUUUUGCUAUUAUAAUUACAGCAUUUGCCGCUGAUGAUGUUGUUUU___ ........(((((..((((....))))........(((.............)))..)))))................... ( -7.54 = -7.51 + -0.04)
| Location | 23,383,039 – 23,383,113 |
|---|---|
| Length | 74 |
| Sequences | 7 |
| Columns | 80 |
| Reading direction | reverse |
| Mean pairwise identity | 70.32 |
| Shannon entropy | 0.58815 |
| G+C content | 0.40381 |
| Mean single sequence MFE | -17.97 |
| Consensus MFE | -8.63 |
| Energy contribution | -8.41 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
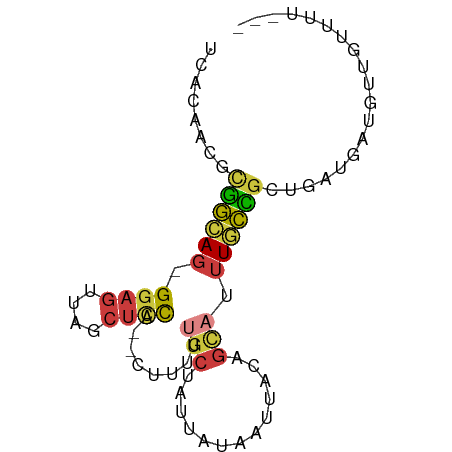
>dm3.chr3R 23383039 74 - 27905053 ---AAAACAACAUUAUCAGCGGCAAAUGCUGUAAUUAUAAUGGCAAAAAG--UGGAGCUAACUCC-CUGCCGCGUUGUGA ---...(((((.......(((((...((((((.......))))))...((--.((((....))))-)))))))))))).. ( -22.21, z-score = -2.55, R) >droSim1.chr3R 23114249 74 - 27517382 ---AAAACAACAUCAUCAGCGGCAAAUGCUGUAAUUAUAAUGGCAAAAAG--UGGAGCUAACUCC-CUGCCGCGUUGUGA ---.........((((..(((((...((((((.......))))))...((--.((((....))))-)))))))...)))) ( -22.70, z-score = -2.69, R) >droSec1.super_22 16858 74 - 1066717 ---AAAACAACAUCAUCAGCGGCAAAUGCUGUAAUUAUAAUGGCAAAAAG--UGGAGCUAACUCC-CUGCCGCGUUGUGA ---.........((((..(((((...((((((.......))))))...((--.((((....))))-)))))))...)))) ( -22.70, z-score = -2.69, R) >droYak2.chr3R 5725795 75 + 28832112 ---AAAACAACAUCAUCAGCGGCAAAUGCUGUAAUUAUAGUAGCAAAAAA-CUGGAGCUAACUCC-CUGCCGAGUUGUGA ---...(((((........(((((..((((((....))))))........-..((((....))))-.))))).))))).. ( -18.20, z-score = -1.74, R) >droEre2.scaffold_4820 5813936 75 + 10470090 ---AAAACAACAUCAUCAGCGGCAAAUGCUGUAAUUAUAGUAGCAAAAAA-CUGGAGCUAACUCC-CUGCCGCGUUGUGA ---.........((((..((((((..((((((....))))))........-..((((....))))-.))))))...)))) ( -21.80, z-score = -2.81, R) >droPer1.super_0 2842567 65 - 11822988 -------------AAACAAACAAAAAAACUGUAAUUAUAAUAGCAACUCC--CGCCGGGGGCUCCGCUUUGAAGUUGUGA -------------.....................((((((((((..((((--(...)))))....))).....))))))) ( -8.80, z-score = 1.09, R) >droWil1.scaffold_181089 3621508 80 - 12369635 ACUCUAUAAUUAUAAUUAUCAGGAGGACGUAUAAUUGUGACAUCAUCCAAAAUAAAGUGAAUUCCAUUGCUAUGUUGUUA .(((((((((....)))))..))))((((((....((.((..((((..........))))..))))....)))))).... ( -9.40, z-score = 0.87, R) >consensus ___AAAACAACAUCAUCAGCGGCAAAUGCUGUAAUUAUAAUAGCAAAAAA__UGGAGCUAACUCC_CUGCCGCGUUGUGA ............(((.((((((((..((((((.......))))))........((((....))))..))))..))))))) ( -8.63 = -8.41 + -0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:48:28 2011