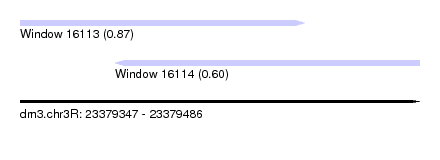
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,379,347 – 23,379,486 |
| Length | 139 |
| Max. P | 0.868450 |

| Location | 23,379,347 – 23,379,446 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 74.47 |
| Shannon entropy | 0.48850 |
| G+C content | 0.44364 |
| Mean single sequence MFE | -21.86 |
| Consensus MFE | -13.30 |
| Energy contribution | -13.80 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
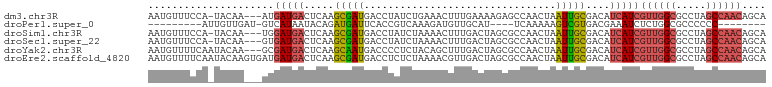
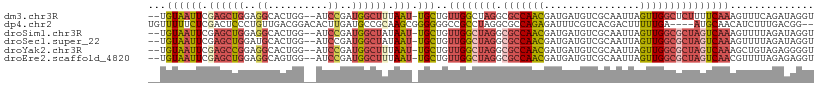
>dm3.chr3R 23379347 99 + 27905053 AAUGUUUCCA-UACAA---AUGAUGACUCAAGCGAUGACCUAUCUGAAACUUUGAAAAGAGCCAACUAAUUGCGACAUCAUCGUUGGCGCCUAGCCAACAGCA ..(((.....-.))).---..(((((.....(((((((....)).....((((....)))).......)))))....)))))((((((.....)))))).... ( -21.40, z-score = -1.55, R) >droPer1.super_0 2838353 81 + 11822988 ---------AUUGUUGAU-GUCAUAAUACAGAUGAUUCACCGUCAAAGAUGUUGCAU----UCAAAAAGUCGUGACGAAAUCUCUGGCGCCCCCC-------- ---------.........-.........((((.((((...(((((..(((.(((...----.)))...))).))))).)))))))).........-------- ( -14.10, z-score = 0.01, R) >droSim1.chr3R 23110508 99 + 27517382 AAUGUUUCCA-UACAA---UGGAUGACUCAAGCGAUGACCUAUCUAAAACUUUGACUAGCGCCAACUAAUUGCGACAUCAUCGUUGGCGCCUAGCCAACAGCA ...(((((((-(...)---)))).)))....(((((((.(((((.........)).)))(((.........)))...)))))((((((.....)))))).)). ( -23.90, z-score = -2.04, R) >droSec1.super_22 13123 99 + 1066717 AAUGUUUCCA-UACAA---GUGAUGACUCAAGCGAUGACCUAUCUAAAACUUUGACUAGCGCCAACUAAUUGCGACAUCAUCGUUGGCGCCUAGCCAACAGCA ...(((..((-(....---)))..)))....(((((((.(((((.........)).)))(((.........)))...)))))((((((.....)))))).)). ( -21.80, z-score = -1.26, R) >droYak2.chr3R 5721973 100 - 28832112 AAUGUUUUCAAUACAA---GCGAUGACUCAAGCAAUGACCCCUCUACAGCUUUGACUAGCGCCAACUAAUUGCGACAUCAUCGUUGGCGCCUAGCCAACAGCA ................---(((((((.....(((((............(((......)))........)))))....)))))((((((.....)))))).)). ( -25.25, z-score = -2.20, R) >droEre2.scaffold_4820 5810058 103 - 10470090 AAUGUUUUCAAUACAAGUGAUGAUGACUCAAGCGAUGACCUCUCUAAAACGUUGACUAGCGCCAACUAAUUGCGACAUCAUCGUUGGCGCCUAGCCAACAGCA ...(((.(((.((....)).))).)))....(((((((..((.(......((((........)))).....).))..)))))((((((.....)))))).)). ( -24.70, z-score = -1.25, R) >consensus AAUGUUUCCA_UACAA___GUGAUGACUCAAGCGAUGACCUAUCUAAAACUUUGACUAGCGCCAACUAAUUGCGACAUCAUCGUUGGCGCCUAGCCAACAGCA .....................(((((.....(((((................................)))))....)))))((((((.....)))))).... (-13.30 = -13.80 + 0.50)
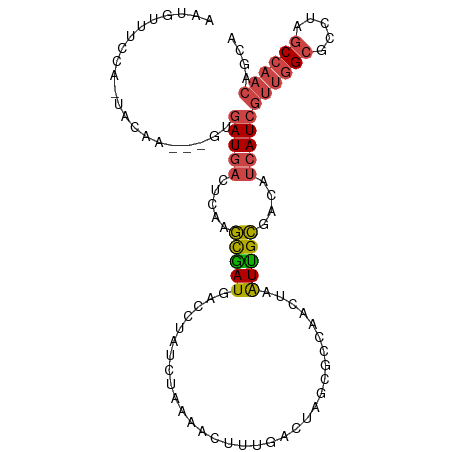
| Location | 23,379,380 – 23,379,486 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.89 |
| Shannon entropy | 0.41573 |
| G+C content | 0.49142 |
| Mean single sequence MFE | -34.89 |
| Consensus MFE | -20.28 |
| Energy contribution | -21.04 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.595236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
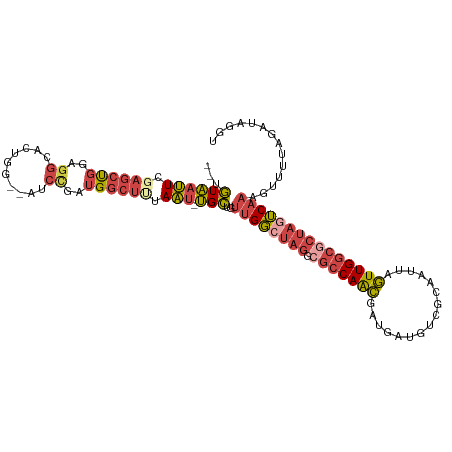
>dm3.chr3R 23379380 106 - 27905053 --UGUAAUUCGAGCUGGAGGCACUGG--AUCCGAUGGCUUUAAU-UGCUGUUGGCUAGGCGCCAACGAUGAUGUCGCAAUUAGUUGGCUCUUUUCAAAGUUUCAGAUAGGU --......((...((((((((.((((--..((((((((......-.))))))))))))))((((((..((((......))))))))))...........))))))...)). ( -30.70, z-score = -0.52, R) >dp4.chr2 9025547 105 - 30794189 UGUUUUUCUCGACUCCCUGUUGACGGACACUUGAUGCCGCAAGCGGGGGGCCGCCUAGGCGCCAGAGAUUUCGUCACGACUUUUUGA----AUGCAACAUCUUUGACGG-- ......((((....(((((((..(((.((.....)))))..)))))))(((.((....))))).))))...(((((.......(((.----...)))......))))).-- ( -32.42, z-score = -0.82, R) >droSim1.chr3R 23110541 106 - 27517382 --UGUAAUUCGAGCUGGAGGCACUGG--AUCCGAUGGCUAUAAU-UGCUGUUGGCUAGGCGCCAACGAUGAUGUCGCAAUUAGUUGGCGCUAGUCAAAGUUUUAGAUAGGU --.((((((..(((((..((......--..))..)))))..)))-)))..((((((((.(((((((..((((......))))))))))))))))))).............. ( -33.90, z-score = -1.36, R) >droSec1.super_22 13156 106 - 1066717 --UGUAAUUCGAGCUGGAUGCACUGG--AUCCGAUGGCUAUAAU-UGCUGUUGGCUAGGCGCCAACGAUGAUGUCGCAAUUAGUUGGCGCUAGUCAAAGUUUUAGAUAGGU --.((((((..((((((((.(...).--))))...))))..)))-)))..((((((((.(((((((..((((......))))))))))))))))))).............. ( -34.20, z-score = -1.57, R) >droYak2.chr3R 5722007 106 + 28832112 --UGUAAUUCGAGCCGGAGGCACUGG--AUCCGAUGGCUUUAAU-UGCUGUUGGCUAGGCGCCAACGAUGAUGUCGCAAUUAGUUGGCGCUAGUCAAAGCUGUAGAGGGGU --.((((((.((((((..((......--..))..)))))).)))-)))..((((((((.(((((((..((((......))))))))))))))))))).............. ( -38.80, z-score = -1.79, R) >droEre2.scaffold_4820 5810095 106 + 10470090 --UGUAAUUCGAGCUGGAGGCAGUGG--AUCCGAUGGCUUUAAU-UGCUGUUGGCUAGGCGCCAACGAUGAUGUCGCAAUUAGUUGGCGCUAGUCAACGUUUUAGAGAGGU --.((((((.((((((..((......--..))..)))))).)))-))).(((((((((.(((((((..((((......))))))))))))))))))))............. ( -39.30, z-score = -2.78, R) >consensus __UGUAAUUCGAGCUGGAGGCACUGG__AUCCGAUGGCUUUAAU_UGCUGUUGGCUAGGCGCCAACGAUGAUGUCGCAAUUAGUUGGCGCUAGUCAAAGUUUUAGAUAGGU ................(((((.........((((((((........))))))))...(((((((((................))))))))).......)))))........ (-20.28 = -21.04 + 0.76)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:48:27 2011