| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,342,352 – 23,342,432 |
| Length | 80 |
| Max. P | 0.759730 |

| Location | 23,342,352 – 23,342,432 |
|---|---|
| Length | 80 |
| Sequences | 6 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 73.57 |
| Shannon entropy | 0.48095 |
| G+C content | 0.44930 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -8.24 |
| Energy contribution | -9.69 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.621752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
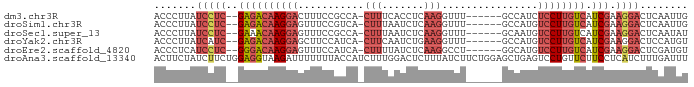
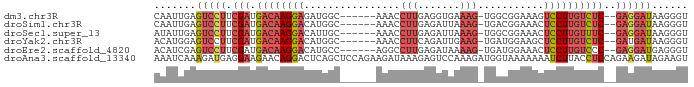
>dm3.chr3R 23342352 80 + 27905053 ACCCUUAUCCUC--GAGACAAGGACUUUCCGCCA-CUUUCACCUCAAGGUUU------GCCAUCUCCUUGUCAUCGAAGGACUCAAUUG .......(((((--((((((((((......((.(-((((......)))))..------))....)))))))).))).))))........ ( -21.90, z-score = -3.56, R) >droSim1.chr3R 23079849 80 + 27517382 ACCCUUAUCCUC--GAGACAAGGAGUUUCCGUCA-CUUUAAUCUCAAGGUUU------GCCAUGUCCUUGUCAUCGAAGGACUCAAUUG .......(((((--((((((((((.........(-((((......)))))..------......)))))))).))).))))........ ( -20.93, z-score = -2.23, R) >droSec1.super_13 2085571 80 + 2104621 ACCCUUAUCCUC--GAAACAAGGAGUUUCCGCCA-CUUUAAUCUCAAGGUUU------GCAAUGUCCUUGUCAUCGAAGGACUCAAUAU .......(((((--((.(((((((......((.(-((((......)))))..------))....)))))))..))).))))........ ( -17.80, z-score = -1.91, R) >droYak2.chr3R 5692372 80 - 28832112 ACCCUUAUCAUC--GAGACAAGGAGCUUCCAUCA-CUUCAAUCUGAAGGUUU------GCCAUGUCCUUGUCAUCGAAGGACUCCAUGU ..(((.....((--((((((((((((..((.(((-........))).))...------))....)))))))).)))))))......... ( -22.40, z-score = -1.99, R) >droEre2.scaffold_4820 5781501 80 - 10470090 ACCCUCAUCCUC--GGGACAAGGAGUUUCCAUCA-CUUUUAUCUCAAGGCCU------GGCAUGUCCUUGUCAUCGAAGGACUCGAUGU .......(((((--((((((((((....(((...-(((.......)))...)------))....)))))))).))).))))........ ( -22.30, z-score = -0.85, R) >droAna3.scaffold_13340 4363288 89 - 23697760 ACUUCUAUCUUCUGGAGGUAAGAUUUUUUUACCAUCUUUGGACUCUUUAUCUUCUGGAGCUGAGUCCUGUUCUUCCUCAUCUUUGAUUU .............((((((((((...)))))))......((((((....((.....))...))))))......)))(((....)))... ( -18.90, z-score = -1.20, R) >consensus ACCCUUAUCCUC__GAGACAAGGAGUUUCCACCA_CUUUAAUCUCAAGGUUU______GCCAUGUCCUUGUCAUCGAAGGACUCAAUUU .......((((...((((((((((...............((((....)))).............)))))))).))..))))........ ( -8.24 = -9.69 + 1.45)
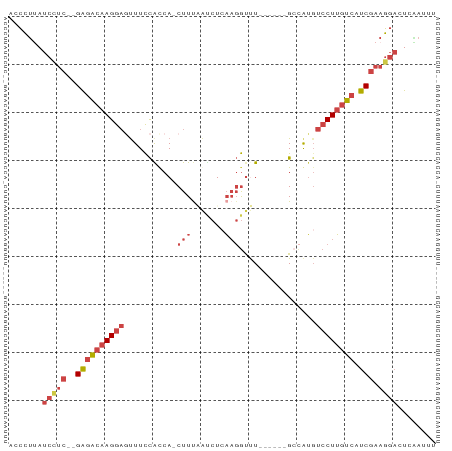
| Location | 23,342,352 – 23,342,432 |
|---|---|
| Length | 80 |
| Sequences | 6 |
| Columns | 89 |
| Reading direction | reverse |
| Mean pairwise identity | 73.57 |
| Shannon entropy | 0.48095 |
| G+C content | 0.44930 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -10.99 |
| Energy contribution | -11.85 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.759730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
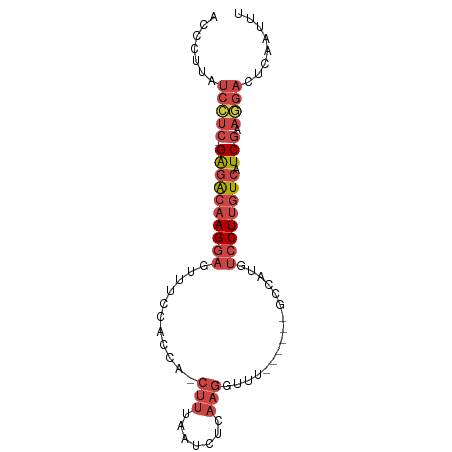
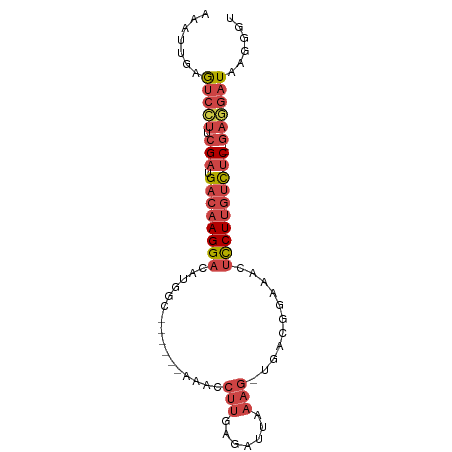
>dm3.chr3R 23342352 80 - 27905053 CAAUUGAGUCCUUCGAUGACAAGGAGAUGGC------AAACCUUGAGGUGAAAG-UGGCGGAAAGUCCUUGUCUC--GAGGAUAAGGGU .......(((((.(((.((((((((..((.(------(.(((....))).....-)).)).....))))))))))--))))))...... ( -26.80, z-score = -2.64, R) >droSim1.chr3R 23079849 80 - 27517382 CAAUUGAGUCCUUCGAUGACAAGGACAUGGC------AAACCUUGAGAUUAAAG-UGACGGAAACUCCUUGUCUC--GAGGAUAAGGGU .......(((((.(((.((((((((((.((.------...)).)).........-....(....)))))))))))--))))))...... ( -25.00, z-score = -2.34, R) >droSec1.super_13 2085571 80 - 2104621 AUAUUGAGUCCUUCGAUGACAAGGACAUUGC------AAACCUUGAGAUUAAAG-UGGCGGAAACUCCUUGUUUC--GAGGAUAAGGGU .......(((((.(((.((((((((....((------....(((.......)))-..))(....)))))))))))--))))))...... ( -22.70, z-score = -1.86, R) >droYak2.chr3R 5692372 80 + 28832112 ACAUGGAGUCCUUCGAUGACAAGGACAUGGC------AAACCUUCAGAUUGAAG-UGAUGGAAGCUCCUUGUCUC--GAUGAUAAGGGU .......(((..((((.((((((((....((------...((.(((........-))).))..))))))))))))--)).)))...... ( -21.40, z-score = -0.42, R) >droEre2.scaffold_4820 5781501 80 + 10470090 ACAUCGAGUCCUUCGAUGACAAGGACAUGCC------AGGCCUUGAGAUAAAAG-UGAUGGAAACUCCUUGUCCC--GAGGAUGAGGGU ...((..(((((.((..((((((((....((------(...(((.......)))-...)))....)))))))).)--))))))..)).. ( -25.90, z-score = -1.73, R) >droAna3.scaffold_13340 4363288 89 + 23697760 AAAUCAAAGAUGAGGAAGAACAGGACUCAGCUCCAGAAGAUAAAGAGUCCAAAGAUGGUAAAAAAAUCUUACCUCCAGAAGAUAGAAGU ..(((......((((.......((((((................)))))).(((((.........))))).)))).....)))...... ( -16.49, z-score = -1.48, R) >consensus AAAUUGAGUCCUUCGAUGACAAGGACAUGGC______AAACCUUGAGAUUAAAG_UGACGGAAACUCCUUGUCUC__GAGGAUAAGGGU .......((((((....((((((((........................................))))))))....))))))...... (-10.99 = -11.85 + 0.86)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:48:23 2011