| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,336,020 – 23,336,103 |
| Length | 83 |
| Max. P | 0.701028 |

| Location | 23,336,020 – 23,336,103 |
|---|---|
| Length | 83 |
| Sequences | 10 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 69.04 |
| Shannon entropy | 0.63155 |
| G+C content | 0.43396 |
| Mean single sequence MFE | -16.45 |
| Consensus MFE | -9.47 |
| Energy contribution | -9.40 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.52 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.701028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23336020 83 - 27905053 GCCGUGACAGAUGGGUGGACAUUUUGUCAGCGAGCUACAAAAAA--UGGGCAAAAAAUGCAAAAAAAAAAGUUGGGGAUCUAGAU------- (((((((((((((......)).))))))).)).)).........--...(((.....))).........................------- ( -13.90, z-score = -0.03, R) >droSim1.chr3R 23073542 82 - 27517382 GCCGUGACAGAUGGGUGGACAUUUUGUCAGCGAGCUACAAAAAA--UGGGCAAAAAAUGCAAAAAAAAGA-UGGGGGAUCUACGU------- .((((.....))))(((((..(((..((((....))........--...(((.....)))........))-..)))..)))))..------- ( -15.30, z-score = -0.31, R) >droSec1.super_13 2079322 82 - 2104621 GCCGUGACAGAUGGGUGGACAUUUUGUCAGCGAGCUACAAAAAA--UGGGCAAAAAUGCAAAAAAAAAGA-UGGGGGAUCUAGAU------- (((((((((((((......)).))))))).)).)).........--...(((....)))........(((-(.....))))....------- ( -15.10, z-score = -0.64, R) >droYak2.chr3R 5685384 80 + 28832112 GCCGUGACAGAUGGGUGGACAUUUUGUCAGCGAGCGACAAAAAA--UGGGCAAAAAAUGC--GAAAAAGA-UGGGCGAUCUAGAU------- (((((((((((((......)).))))))).)).)).........--...(((.....)))--.....(((-(.....))))....------- ( -15.60, z-score = -0.09, R) >droEre2.scaffold_4820 5775160 80 + 10470090 GCCGUGACAGAUGGGUGGACAUUUUGUCAGCGAACGACAAAAAC--UGGGCAAAAAAUGC--GCAAAAGA-UGGGGGAUCUAGAU------- ..(((((((((((......)).))))))).))............--((.(((.....)))--.))..(((-(.....))))....------- ( -14.90, z-score = 0.15, R) >droAna3.scaffold_13340 4356925 90 + 23697760 GUCGUGACAGAUGGGUGGACAUUUUGUCAGCGAGUGC-AGAUAGUCCAAAAAGGAGUUGC-AAAAAAUUUCAAAAGGAUUAAGAUCAUUGAU .((((((((((((......)).))))))).))).(((-((....(((.....))).))))-).......((((...(((....))).)))). ( -19.30, z-score = -1.18, R) >dp4.chr2 8995401 82 - 30794189 GCCGUGACAGAUGGGUGGACAUUUUGUCAGCGUGCAUGAGCAAGAGUCGGCAAAAAACGC-AAGAAACAUUAAGUUUAGCCAA--------- ...........(((.(((((.(((((((.((.(((....)))...)).)))))))..(..-..).........))))).))).--------- ( -21.50, z-score = -1.11, R) >droPer1.super_0 2808212 82 - 11822988 GCCGUGACAGAUGGGUGGACAUUUUGUCAGCGUGCAUGAGCAAGAGUCGGCAAAAAACGC-AAGAAACAUUAAGUUUAGCCAA--------- ...........(((.(((((.(((((((.((.(((....)))...)).)))))))..(..-..).........))))).))).--------- ( -21.50, z-score = -1.11, R) >droWil1.scaffold_181089 3581400 71 - 12369635 GCUGUGACAGAUGGGUGGACAUUCUGUCAGCAAGUGC-AG-------CAAAAGAGGAUGC-AGGACAC--CCAAAGGAUCUA---------- (((.(((((((((......)).)))))))((....))-))-------)......((((.(-.((....--))...).)))).---------- ( -18.40, z-score = -0.08, R) >droGri2.scaffold_15074 6747674 79 - 7742996 GCAGUGACAGAUGGGUGGACAUUUUGUCAGAGAG---UAUAAAG--UGAAAAACAGUUACAAACAAAAAAAUAAAAUAUAUAGA-------- ...(((((...((..(...(((((((((.....)---.))))))--))..)..)))))))........................-------- ( -9.00, z-score = -0.80, R) >consensus GCCGUGACAGAUGGGUGGACAUUUUGUCAGCGAGCGACAAAAAA__UGGGCAAAAAAUGC_AAAAAAAAA_UAGGGGAUCUAGAU_______ (((((((((((((......)).))))))).)).))......................................................... ( -9.47 = -9.40 + -0.07)

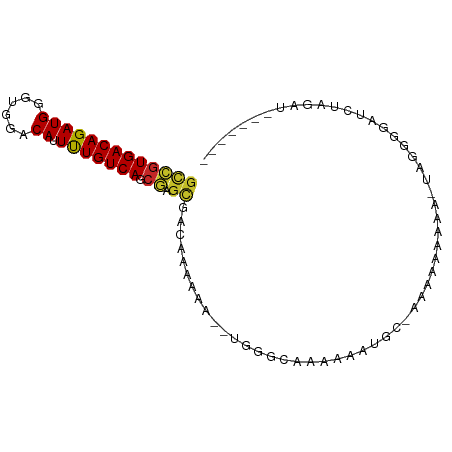
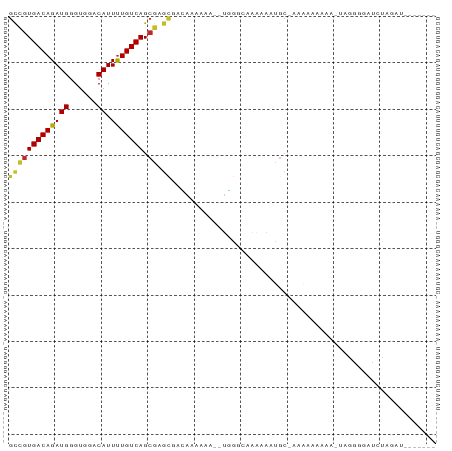
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:48:21 2011