
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,325,791 – 23,325,882 |
| Length | 91 |
| Max. P | 0.696641 |

| Location | 23,325,791 – 23,325,882 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 74.39 |
| Shannon entropy | 0.50697 |
| G+C content | 0.41112 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -11.46 |
| Energy contribution | -10.66 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.696641 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
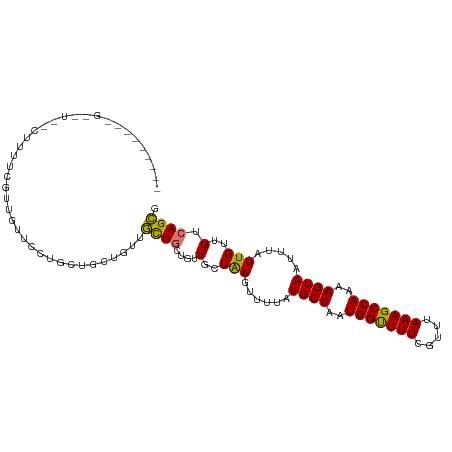
>dm3.chr3R 23325791 91 - 27905053 ---------------GUUGGCU-GGUUGCGGCCGCUGUUGUUGCUGUUGCCAAGUUUUAUGCUAAUGGUUUUCGUUUAAAGCCAAAAGUAAUUUAUUGUUGUCAGCG ---------------(((((((-(((.(((((.((....)).))))).)))).......((((..(((((((.....)))))))..))))..........)))))). ( -29.40, z-score = -2.96, R) >droEre2.scaffold_4820 5764947 91 + 10470090 ---------------GUUGGCU-GGUUGCUGCUGCUGUUGUUGCUGUUGCCAAGUUUUAUGCUAAUGGUUUUCGUUUAAAGCCAAAAGUAAUUUAUUGUUGUCAGCG ---------------(((((((-(((.((.((.((....)).)).)).)))).......((((..(((((((.....)))))))..))))..........)))))). ( -25.50, z-score = -2.06, R) >droAna3.scaffold_13340 4346525 106 + 23697760 CCUGGUCCUGGUCCUAUACUCG-UGUUGCGGCCGCUGUUGUUGCUGUUGCCAAGUUUUAUGCUAAUGGUUUUCGUUUAAAGCCAAAAGUAAUUUAUUGUUGUCAGCG .((((...((((.....(((.(-.((.(((((.((....)).))))).))).))).....)))).(((((((.....))))))).................)))).. ( -20.90, z-score = 0.38, R) >dp4.chr2 8984807 101 - 30794189 ------CUCGUGUUCCCUUCCGUUCGUCGUUUUGUUGUUGCUCCUGUUGCCAAGUUUUAUGCUAAUGGUUUUCGUUUAAAGCCAAAAGUAAUUUAUUGUUGUCAGCG ------...........................((((..((....(((((..(((.....)))..(((((((.....)))))))...))))).....))...)))). ( -13.40, z-score = -0.63, R) >droPer1.super_0 2798017 101 - 11822988 ------CUCGUGUUCCCUUCCGUUCGUUGUUUUGUUGUUGCUCCUGUUGCCAAGUUUUAUGCUAAUGGUUUUCGUUUAAAGCCAAAAGUAAUUUAUUGUUGUCAGCG ------...........................((((..((....(((((..(((.....)))..(((((((.....)))))))...))))).....))...)))). ( -13.40, z-score = -0.62, R) >droMoj3.scaffold_6540 13808351 84 - 34148556 --------------------GGCUGCUCCUCUUCCA---ACUGAUGAUGCCAAGUUUUAUGCUAAUGGCUUUCGUUUAAAGCCAAAAGUAAUUUAUUGUUGUCAGUG --------------------................---(((((..(((..((((....((((..(((((((.....)))))))..))))))))..)))..))))). ( -18.60, z-score = -1.55, R) >droVir3.scaffold_13047 1369291 97 + 19223366 ------GCUGGUUGGCU-UUUGUUGUCCCUACUCCU---ACUGAUGAUGCCGAGUUUUAUGCUAAUGGCUUUCGUUUAAAGCCAAAAGUAAUUUAUUGUUGUCAGUG ------...((.((((.-...)))).)).......(---(((((..(((..((((....((((..(((((((.....)))))))..))))))))..)))..)))))) ( -21.20, z-score = -1.30, R) >droGri2.scaffold_15074 6735423 104 - 7742996 ---GCUGCUAGCUGGUUGUUCGUUGUUCCUACUCCUGCUACUGAUGAUGCCAAGUUUUAUGCUAAUGGCUUUCGCUUAAAGCCAAAAGUAAUUUAUUGUUGUCAGUG ---((((.((((((((...(((((((..(.......)..)).))))).)))).......((((..(((((((.....)))))))..)))).......)))).)))). ( -24.00, z-score = -1.13, R) >consensus _________G__U__CUUUUCGUUGUUCCUGCUGCUGUUGCUGCUGUUGCCAAGUUUUAUGCUAAUGGUUUUCGUUUAAAGCCAAAAGUAAUUUAUUGUUGUCAGCG .......................................((((....(..(((......((((..(((((((.....)))))))..)))).....)))..).)))). (-11.46 = -10.66 + -0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:48:18 2011