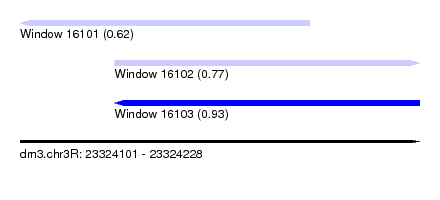
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,324,101 – 23,324,228 |
| Length | 127 |
| Max. P | 0.932018 |

| Location | 23,324,101 – 23,324,193 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 66.70 |
| Shannon entropy | 0.64153 |
| G+C content | 0.50194 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -12.51 |
| Energy contribution | -12.01 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.619548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
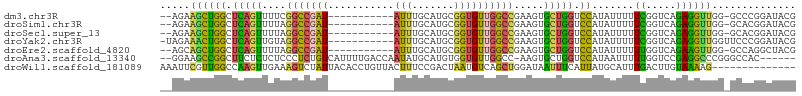
>dm3.chr3R 23324101 92 - 27905053 --AGAAGCUGGCUCAGUUUUCGGCCGAU-----------AUUUGCAUGCGGUGUUGGCCGAAGUGCUGGUCCAUAUUUUUCGGUCAGAGGUUGG-GCCCGGAUACG --.....((((..(((((((((((((((-----------(((.(....))))))))))))))).))))(((((....((((.....)))).)))-))))))..... ( -33.70, z-score = -1.58, R) >droSim1.chr3R 23061616 92 - 27517382 --AGAAGCUGGCUCAGUUUUAGGCCGAU-----------AUUUGCAUGCGGUGUUGGCCGAAGUGCUGGUCCAUAUUUUUCGGUCAGAGGUUGG-GCACGGAUACG --.(((((((...))))))).(((((((-----------(((.(....)))))))))))...(((((((((((....((((.....)))).)))-)).))).))). ( -30.60, z-score = -1.31, R) >droSec1.super_13 2067407 92 - 2104621 --AGAAGCUGGCUCAGUUUUAGGCCGAU-----------AUUUGCAUGCGGUGUUGGCCGAAGUGCUGGUCCAUAUUUUUCGGUCAGAGGUUGG-GCACGGAUACG --.(((((((...))))))).(((((((-----------(((.(....)))))))))))...(((((((((((....((((.....)))).)))-)).))).))). ( -30.60, z-score = -1.31, R) >droYak2.chr3R 5673010 94 + 28832112 -UAGAAACUGGCUCAGUUGUAGGCCGAU-----------AUUUGCAUGCGGUGUUGGCCGAAGUGCUGGUCCAUAUUUUUCGGUCAGAGGUUGGUUCCCGGAUACG -..((((.(((..((((....(((((((-----------(((.(....))))))))))).....))))..)))....)))).(((.(.((......))).)))... ( -26.50, z-score = 0.03, R) >droEre2.scaffold_4820 5763231 92 + 10470090 --AGCAGCUGGCUCAGUUUUAGGCCGAU-----------AUUUGCAUGCGGUGUUGGCCGAAGUGCUGGUCCAUAUUUUUUGGUCAGAAGUUGG-GCCAGGCUACG --(((..((((((((..((..(((((((-----------(((.(....)))))))))))......((((.(((.......)))))))))..)))-))))))))... ( -35.50, z-score = -2.24, R) >droAna3.scaffold_13340 4344468 97 + 23697760 --GGAAGCCGGCUUCUCUCUCCCUCUGUCAUUUUGACCAAUAUGCAUGUGGUGUUGGCC-AAGUGCUGGUCCAUAAUUUUUGGUCCGAGGCCCGGGCCAC------ --((...((((((((...........(((.....)))......((((.((((....)))-).)))).((.(((.......))).)))))).)))).))..------ ( -30.50, z-score = -0.39, R) >droWil1.scaffold_181089 3566432 92 - 12369635 AAAUUCGUUGGCCAAGUUGAAAGUCUAUUACACCUGUUACUUUCCGACUAAUGUCAGCUGGAUAAUUUCAUUAUGCAUUUGACUUGUAAAAG-------------- ..((((((((((..((((((((((.....((....)).))))).)))))...)))))).))))..........((((.......))))....-------------- ( -16.00, z-score = -0.61, R) >consensus __AGAAGCUGGCUCAGUUUUAGGCCGAU___________AUUUGCAUGCGGUGUUGGCCGAAGUGCUGGUCCAUAUUUUUCGGUCAGAGGUUGG_GCCCGGAUACG .....((((((((........))))............................((((((((((..............)))))))))).)))).............. (-12.51 = -12.01 + -0.50)
| Location | 23,324,131 – 23,324,228 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 70.51 |
| Shannon entropy | 0.56266 |
| G+C content | 0.48373 |
| Mean single sequence MFE | -25.91 |
| Consensus MFE | -14.95 |
| Energy contribution | -15.61 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.766727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
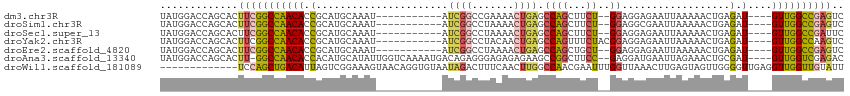
>dm3.chr3R 23324131 97 + 27905053 UAUGGACCAGCACUUCGGCCAACACCGCAUGCAAAU-----------AUCGGCCGAAAACUGAGCCAGCUUCU--GGAGGAGAAUUAAAAACUGAGAU----GUUGGCCGAGUC .............(((((((((((.(.((.......-----------...(((((.....)).)))..((((.--...))))..........)).).)----)))))))))).. ( -26.40, z-score = -1.02, R) >droSim1.chr3R 23061646 97 + 27517382 UAUGGACCAGCACUUCGGCCAACACCGCAUGCAAAU-----------AUCGGCCUAAAACUGAGCCAGCUUCU--GGAGGCGAAUUAAAAACUGAGAU----GUUGGCCGAGUC .............(((((((((((.(.((.......-----------...((((.......).))).((((..--..))))...........)).).)----)))))))))).. ( -27.50, z-score = -1.20, R) >droSec1.super_13 2067437 97 + 2104621 UAUGGACCAGCACUUCGGCCAACACCGCAUGCAAAU-----------AUCGGCCUAAAACUGAGCCAGCUUCU--GGAGGAGAAUUAAAAACUGAGAU----GUUGGCCGAUUC ..............((((((((((.(.((.......-----------...((((.......).)))..((((.--...))))..........)).).)----)))))))))... ( -25.70, z-score = -1.25, R) >droYak2.chr3R 5673041 99 - 28832112 UAUGGACCAGCACUUCGGCCAACACCGCAUGCAAAU-----------AUCGGCCUACAACUGAGCCAGUUUCUACGGAGGAGAAUUAAAAACUGAGAU----GUUGGCCAAGUC ...........((((.((((((((.(.((.......-----------...((((.......).)))..((((....))))............)).).)----))))))))))). ( -24.40, z-score = -0.73, R) >droEre2.scaffold_4820 5763261 97 - 10470090 UAUGGACCAGCACUUCGGCCAACACCGCAUGCAAAU-----------AUCGGCCUAAAACUGAGCCAGCUGCU--GGAGGAGAAUUAAAAACUGAGAU----GUUGGCCGAGUC .............((((((((((((((((.((....-----------.((((.......))))....))))).--))((............))....)----)))))))))).. ( -26.30, z-score = -0.87, R) >droAna3.scaffold_13340 4344493 107 - 23697760 UAUGGACCAGCACUU-GGCCAACACCACAUGCAUAUUGGUCAAAAUGACAGAGGGAGAGAGAAGCCGGCUUCC--GAGGAUGAAUUAGAAACUGCGAU----GUUGGUCGAGAC ............(((-((((((((.(.(((.(......(((.....)))...(((((...(....)..)))))--..).))).....(......)).)----)))))))))).. ( -26.60, z-score = -0.73, R) >droWil1.scaffold_181089 3566462 101 + 12369635 -------------UCCAGCUGACAUUAGUCGGAAAGUAACAGGUGUAAUAGACUUUCAACUUGGCCAACGAAUUUGGUUAAACUUGAGUAGUUGGGGUUGAGGUUGGUUGUAUU -------------..((((..((.(((..(.((((((.((....)).....)))))).......(((((..(((..(......)..))).))))))..))).))..)))).... ( -24.50, z-score = -0.70, R) >consensus UAUGGACCAGCACUUCGGCCAACACCGCAUGCAAAU___________AUCGGCCUAAAACUGAGCCAGCUUCU__GGAGGAGAAUUAAAAACUGAGAU____GUUGGCCGAGUC .............((((((((((...........................(((..........)))..((((....))))......................)))))))))).. (-14.95 = -15.61 + 0.67)
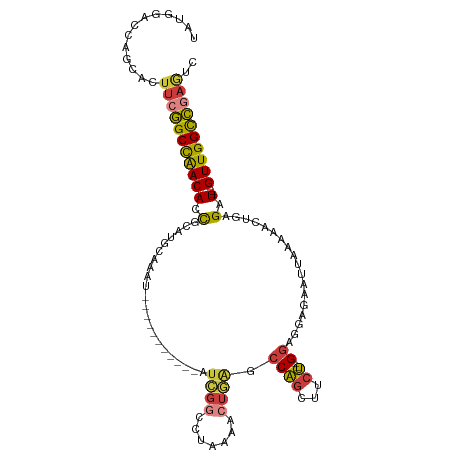
| Location | 23,324,131 – 23,324,228 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 70.51 |
| Shannon entropy | 0.56266 |
| G+C content | 0.48373 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -15.27 |
| Energy contribution | -15.73 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
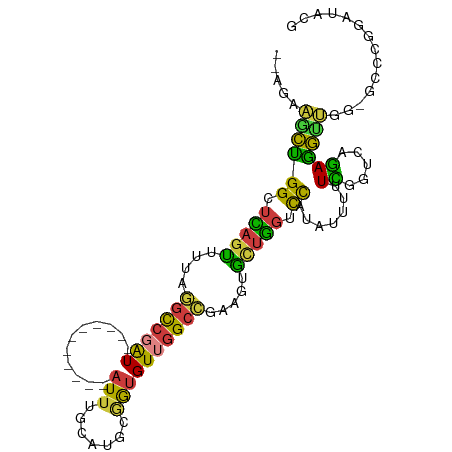
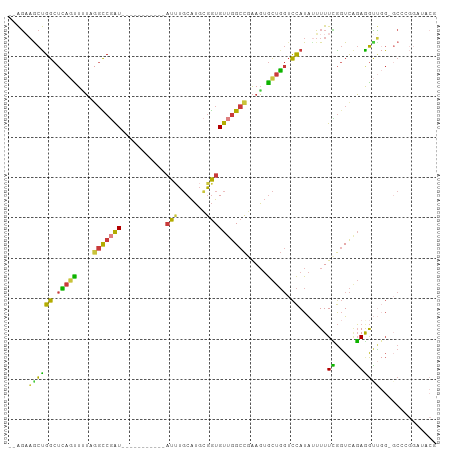
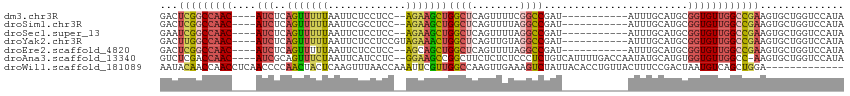
>dm3.chr3R 23324131 97 - 27905053 GACUCGGCCAAC----AUCUCAGUUUUUAAUUCUCCUCC--AGAAGCUGGCUCAGUUUUCGGCCGAU-----------AUUUGCAUGCGGUGUUGGCCGAAGUGCUGGUCCAUA .(((((((((((----(((.((((....(((..((..((--..((((((...))))))..))..)).-----------))).)).)).))))))))))).)))........... ( -31.70, z-score = -1.99, R) >droSim1.chr3R 23061646 97 - 27517382 GACUCGGCCAAC----AUCUCAGUUUUUAAUUCGCCUCC--AGAAGCUGGCUCAGUUUUAGGCCGAU-----------AUUUGCAUGCGGUGUUGGCCGAAGUGCUGGUCCAUA .(((((((((((----(((.((((.....(((.((((..--.(((((((...))))))))))).)))-----------....)).)).))))))))))).)))........... ( -35.10, z-score = -2.69, R) >droSec1.super_13 2067437 97 - 2104621 GAAUCGGCCAAC----AUCUCAGUUUUUAAUUCUCCUCC--AGAAGCUGGCUCAGUUUUAGGCCGAU-----------AUUUGCAUGCGGUGUUGGCCGAAGUGCUGGUCCAUA ...(((((((((----(((.((((....(((..((..((--.(((((((...))))))).))..)).-----------))).)).)).)))))))))))).............. ( -31.00, z-score = -1.88, R) >droYak2.chr3R 5673041 99 + 28832112 GACUUGGCCAAC----AUCUCAGUUUUUAAUUCUCCUCCGUAGAAACUGGCUCAGUUGUAGGCCGAU-----------AUUUGCAUGCGGUGUUGGCCGAAGUGCUGGUCCAUA .(((((((((((----(((.(((((((((...........)))))))))((.....((((((.....-----------.)))))).)))))))))))).))))........... ( -30.90, z-score = -1.67, R) >droEre2.scaffold_4820 5763261 97 + 10470090 GACUCGGCCAAC----AUCUCAGUUUUUAAUUCUCCUCC--AGCAGCUGGCUCAGUUUUAGGCCGAU-----------AUUUGCAUGCGGUGUUGGCCGAAGUGCUGGUCCAUA .(((((((((((----(((..((.........)).....--.((((((((((........)))))..-----------....)).)))))))))))))).)))........... ( -29.10, z-score = -0.99, R) >droAna3.scaffold_13340 4344493 107 + 23697760 GUCUCGACCAAC----AUCGCAGUUUCUAAUUCAUCCUC--GGAAGCCGGCUUCUCUCUCCCUCUGUCAUUUUGACCAAUAUGCAUGUGGUGUUGGCC-AAGUGCUGGUCCAUA .....(((((((----((((((((...............--(((((....)))))..........(((.....)))......)).)))))))))(((.-....))))))).... ( -23.60, z-score = -0.17, R) >droWil1.scaffold_181089 3566462 101 - 12369635 AAUACAACCAACCUCAACCCCAACUACUCAAGUUUAACCAAAUUCGUUGGCCAAGUUGAAAGUCUAUUACACCUGUUACUUUCCGACUAAUGUCAGCUGGA------------- .......(((...........((((.....))))...........((((((..((((((((((.....((....)).))))).)))))...))))))))).------------- ( -15.90, z-score = -0.80, R) >consensus GACUCGGCCAAC____AUCUCAGUUUUUAAUUCUCCUCC__AGAAGCUGGCUCAGUUUUAGGCCGAU___________AUUUGCAUGCGGUGUUGGCCGAAGUGCUGGUCCAUA ...(((((((((.......(((((((((.............)))))))))...........((((......................))))))))))))).............. (-15.27 = -15.73 + 0.46)
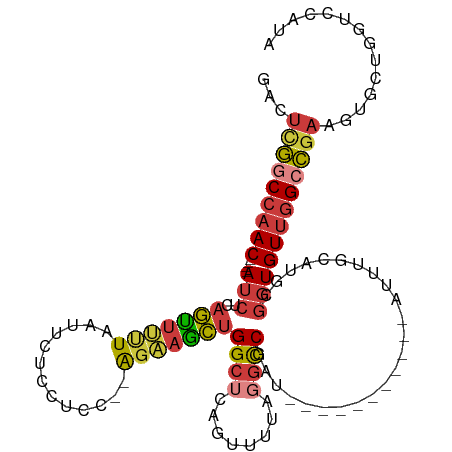
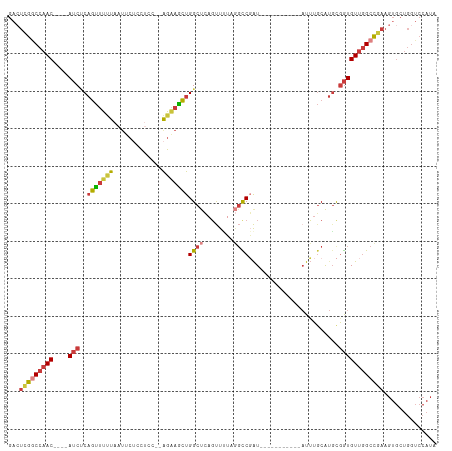
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:48:17 2011