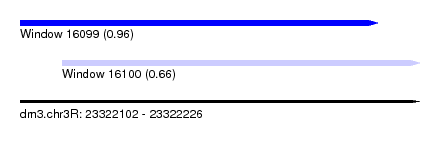
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,322,102 – 23,322,226 |
| Length | 124 |
| Max. P | 0.960400 |

| Location | 23,322,102 – 23,322,213 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.07 |
| Shannon entropy | 0.36144 |
| G+C content | 0.38853 |
| Mean single sequence MFE | -26.53 |
| Consensus MFE | -16.44 |
| Energy contribution | -17.67 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
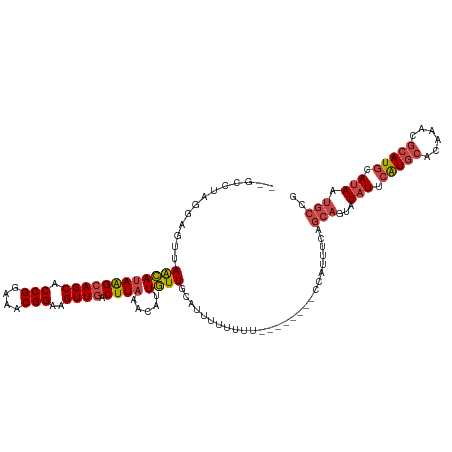
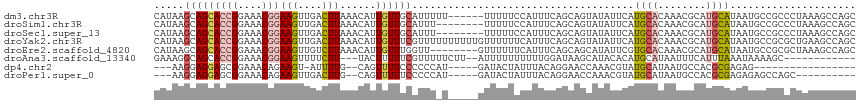
>dm3.chr3R 23322102 111 + 27905053 --GCCUAGGAGUUAACAUAAGCAGCACCGGAAACGGAAGUUGACUUAAACAUUGUUGCAUUUUUUUUUU------CCAUUUCAGCAGUAUAUUCAUGCACAAACGCAUGCAUAAUGCCG --.....((((..((.....(((((((((....)))..(((......)))..)))))).....))..))------))......(((...(((.(((((......))))).))).))).. ( -29.10, z-score = -2.96, R) >droSim1.chr3R 23059666 109 + 27517382 --GCCUAGGAGUUAACAUAAGCAGCACCGGAAACGGAAGUUGACUUAAACAUUGUUGCAUUUUUUUU--------CCAUUUCAGCAGUAUAUUCAUGCACAAACGCAUGCAUAAUGCCG --.....((((..((.((..(((((((((....)))..(((......)))..)))))))).))..))--------))......(((...(((.(((((......))))).))).))).. ( -28.40, z-score = -2.70, R) >droSec1.super_13 2065500 109 + 2104621 --GCCUAGGAGUUAACAUAAGCAGCACCGGAAACGGAAGUUGACUUAAACAUUGUUGCAUUUUUUUU--------CCAUUUCAGCAGUAUAUUCAUGCACAAACGCAUGCAUAAUGCCG --.....((((..((.((..(((((((((....)))..(((......)))..)))))))).))..))--------))......(((...(((.(((((......))))).))).))).. ( -28.40, z-score = -2.70, R) >droYak2.chr3R 5670836 119 - 28832112 CAGCCUAAGAGUUAGCAUAAGCAGCACCGGAAACGGAAGUUGACUUAAACAUUGUUUCGUUUUUUUUUUGUUUUUUCAUUUCAGCAGUAUAUUCAUGCACAAACGCAUGCAUAAUGCCG ........(((..((((((((((((.(((....)))..)))).))))(((........))).......))))..)))......(((...(((.(((((......))))).))).))).. ( -26.40, z-score = -1.57, R) >droEre2.scaffold_4820 5761192 111 - 10470090 CAGCCUAAGAGUUAGCAUAAGCAGCACCGGAAACGGAAGUUGUCUUAAACAUUGUUUGGUUGUUUUU--------UCAUUUCAGCAGCAUAUUCGUGCACAAACGCAUGCAUAAUGCCG (((((.((.(((.....((((((((.(((....)))..)))).))))...))).)).))))).....--------...........((((((.(((((......))))).)).)))).. ( -28.40, z-score = -1.31, R) >droAna3.scaffold_13340 4342360 89 - 23697760 -------------AGGAAAGGCAGCACCGGAAACGGAAGUUUUCUU---UACUUUUUCGUUUUUCUU----------AUUUUU----UUUUUGGAUAAGCAUACACAUGCAUAAUUUCA -------------(((((((((....(((....)))((((......---.))))....)))))))))----------......----...........((((....))))......... ( -18.50, z-score = -2.99, R) >consensus __GCCUAGGAGUUAACAUAAGCAGCACCGGAAACGGAAGUUGACUUAAACAUUGUUGCAUUUUUUUU________CCAUUUCAGCAGUAUAUUCAUGCACAAACGCAUGCAUAAUGCCG .................((((((((.(((....)))..)))).))))....................................(((...(((.(((((......))))).))).))).. (-16.44 = -17.67 + 1.22)

| Location | 23,322,115 – 23,322,226 |
|---|---|
| Length | 111 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 65.12 |
| Shannon entropy | 0.66244 |
| G+C content | 0.41552 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -9.10 |
| Energy contribution | -9.67 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.664019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23322115 111 + 27905053 CAUAAGCAGCACCGGAAACGGAAGUUGACUUAAACAUUGUUGCAUUUUU------UUUUUCCAUUUCAGCAGUAUAUUCAUGCACAAACGCAUGCAUAAUGCCGCCCUAAAGCCAGC .....(((((((((....)))..(((......)))..))))))......------.............((.((((((.(((((......))))).)).)))).))............ ( -27.20, z-score = -2.17, R) >droSim1.chr3R 23059679 109 + 27517382 CAUAAGCAGCACCGGAAACGGAAGUUGACUUAAACAUUGUUGCAUUU--------UUUUUCCAUUUCAGCAGUAUAUUCAUGCACAAACGCAUGCAUAAUGCCGCCCUAAAGCCAGC .....(((((((((....)))..(((......)))..))))))....--------.............((.((((((.(((((......))))).)).)))).))............ ( -27.20, z-score = -2.16, R) >droSec1.super_13 2065513 109 + 2104621 CAUAAGCAGCACCGGAAACGGAAGUUGACUUAAACAUUGUUGCAUUU--------UUUUUCCAUUUCAGCAGUAUAUUCAUGCACAAACGCAUGCAUAAUGCCGCCCUAAAGCCAGC .....(((((((((....)))..(((......)))..))))))....--------.............((.((((((.(((((......))))).)).)))).))............ ( -27.20, z-score = -2.16, R) >droYak2.chr3R 5670851 117 - 28832112 CAUAAGCAGCACCGGAAACGGAAGUUGACUUAAACAUUGUUUCGUUUUUUUUUUGUUUUUUCAUUUCAGCAGUAUAUUCAUGCACAAACGCAUGCAUAAUGCCGCGCUGAAGCCAGC ..((((((((.(((....)))..)))).))))...............................(((((((.((((((.(((((......))))).)).))))...)))))))..... ( -28.90, z-score = -1.50, R) >droEre2.scaffold_4820 5761207 109 - 10470090 CAUAAGCAGCACCGGAAACGGAAGUUGUCUUAAACAUUGUUUGGUU--------GUUUUUUCAUUUCAGCAGCAUAUUCGUGCACAAACGCAUGCAUAAUGCCGCGCUAAAGCCAGC ..((((((((.(((....)))..)))).))))........((((((--------(......)......((.((((((.(((((......))))).)).)))).)).....)))))). ( -29.30, z-score = -1.05, R) >droAna3.scaffold_13340 4342362 100 - 23697760 GAAAGGCAGCACCGGAAACGGAAGUUUUCUU---UACUUUUUCGUUUUUCUU--AUUUUUUUUUUGGAUAAGCAUACACAUGCAUAAUUUCAUUUAAAUAAAAGC------------ (((((((....(((....)))((((......---.))))....)))))))((--((((......((((...((((....)))).....))))...))))))....------------ ( -18.10, z-score = -2.07, R) >dp4.chr2 8981274 89 + 30794189 ---AAGGAGGAGCGGAAACAGAAGU-AUUUUG--CAGUUUUCCCCCCAU-----GAUACUAUUUACAGGAACCAAACGUAUGCAUAAUGCCACGCGAGAG----------------- ---........(((....)....((-(((.((--((((((....((..(-----((......)))..))....))))...)))).)))))...)).....----------------- ( -14.00, z-score = 0.37, R) >droPer1.super_0 2794513 97 + 11822988 ---AAGGAGGAGCGGAAACAGAAGUUGACUUG--CAGUUUUUCCCCCAU-----GAUACUAUUUACAGGAACCAAACGUAUGCAUAAUGCCACGCGAGAGAGCCAGC---------- ---.....((..((....).........((((--(.((((((((....(-----((......)))..))))..))))((((.....))))...))))).)..))...---------- ( -17.30, z-score = 0.32, R) >consensus CAUAAGCAGCACCGGAAACGGAAGUUGACUUAAACAUUGUUUCAUUU_U_____GUUUUUCCAUUUCAGCAGCAUAUUCAUGCACAAACGCAUGCAUAAUGCCGCCCUAAAGCCAGC .....(((((((((....)))(((....)))......)))))).....................................((((........))))..................... ( -9.10 = -9.67 + 0.58)
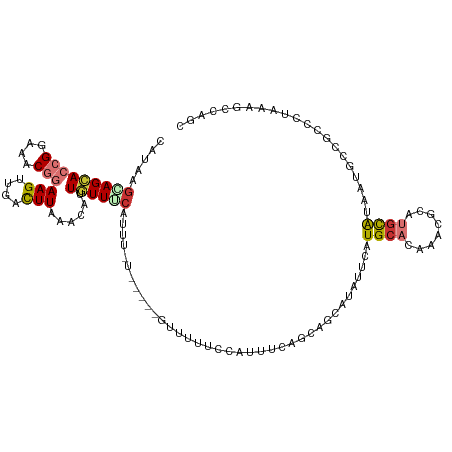
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:48:15 2011