| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,807,528 – 8,807,621 |
| Length | 93 |
| Max. P | 0.776825 |

| Location | 8,807,528 – 8,807,621 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 72.55 |
| Shannon entropy | 0.51099 |
| G+C content | 0.38136 |
| Mean single sequence MFE | -23.09 |
| Consensus MFE | -8.37 |
| Energy contribution | -8.12 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.776825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8807528 93 - 23011544 ---GUCUUCGAAUCGUAAAAUAUGGAAUUGCGUGCAAGUGUUCAAUUAUGUUCU--------CGGAUAU----AUUUCGCUACACUUCACGCAUAUAAUGCGGUCAGU---------- ---......((.(((((...((((....((((((.((((((.....((((((..--------..)))))----).......)))))))))))))))).)))))))...---------- ( -22.90, z-score = -1.80, R) >droSim1.chr2L 8585813 93 - 22036055 ---GUCUUCGAAUCGUAAAAUAUGGAAUUGCGUGCAAGUGUUCAAUUAUGUUCU--------CGGAUAU----AUUUCGCUACACUUCACGCAUAUAAUGCGGUCAGU---------- ---......((.(((((...((((....((((((.((((((.....((((((..--------..)))))----).......)))))))))))))))).)))))))...---------- ( -22.90, z-score = -1.80, R) >droSec1.super_3 4279668 93 - 7220098 ---GUCUUCGAAUCGUAAAAUAUGGAAUUGCGUGCAAGUGUUCAAUUAUGUUCU--------CGGGUAU----AUUUCGCUACACUUCACGCAUAUAAUGCGGUCAGU---------- ---......((.(((((...((((....((((((.((((((......((((...--------.....))----))......)))))))))))))))).)))))))...---------- ( -20.40, z-score = -0.97, R) >droYak2.chr2L 11452332 101 - 22324452 ---GUCUUCGAAUCGUAAAAUAUGGAAUUGCGUGCAAGUGUUCAAUUAUGUUCUCG----GAUAUAUAUGCGCAUUUCGCCACACUUCACGCAUUUAAUGCGGUCAGU---------- ---......((.(((((.......((((.(((((.((((((.....((((((....----))))))...(((.....))).)))))))))))))))..)))))))...---------- ( -23.30, z-score = -0.58, R) >droEre2.scaffold_4929 9399222 105 - 26641161 ---GUCUUCGAAUCGUAAAAUAUGGAAUUGCGUGCAAGUGUUCAAUUAUGUUCUCGCAUAUACGUGUAUGCGCAUUUCGCCACACUUCACGCAUUUAAUGCGGUCAGU---------- ---......((.(((((.......((((.(((((.((((((......(((....(((((((....))))))))))......)))))))))))))))..)))))))...---------- ( -26.80, z-score = -1.03, R) >droAna3.scaffold_12916 10470503 96 - 16180835 ---GUCUUCGAAUCGUAAAAUAUGGAAUUGCGUGCAAGUGUUCAAUUAUGUGC---------CACAUACGCGCAUUUCGCCACACUUCACGCAUUUCAGUCGGUCAGU---------- ---....((((..((((...))))(((.((((((.((((((......((((((---------.......))))))......)))))))))))).)))..)))).....---------- ( -28.00, z-score = -2.64, R) >dp4.chr4_group5 1598407 115 + 2436548 GUCUCUCCCAAAUCGUAAAAUAUGAAAUUGCGUGCAAGUGUUCAAUUAUGUAUCU---CGGAUACGAGUAUAUAUUUCGUUACACUUCACGCAUUUCUUUCGGUCAGCGGUCCCCUCU ...........(((((......(((((.((((((.((((((......((((((((---((....)))).))))))......))))))))))))....)))))....)))))....... ( -31.30, z-score = -4.69, R) >droPer1.super_5 1568166 115 + 6813705 GUCUCUCCCAAAUCGUAAAAUAUGAAAUUGCGUGCAAGUGUUCAAUUAUGUCUCU---CGGAUACGAGUAUAUAUUUCGUUACACUUCACGCAUUUCUUUCGGUCAGCGGUCCCCUCU ...........(((((......(((((.((((((.((((((.....(((((..((---((....)))).))))).......))))))))))))....)))))....)))))....... ( -28.50, z-score = -3.81, R) >droWil1.scaffold_180772 8785312 88 + 8906247 ---GUCUUUAAGUCGUAAAAUAUGAAAUUGCGUGCAAGUGUUAAAUUAUCCUC-----------AGUU-----AUUUCGUUACACUUCACGCAUUUUGC-GGUCCUGC---------- ---.........((((((((........((((((.((((((............-----------....-----........))))))))))))))))))-))......---------- ( -18.36, z-score = -1.92, R) >droGri2.scaffold_15126 7802078 86 - 8399593 ---AUCUUUAAAUCGUAAAAUAUGAAAUUGCGUGCAAGUGUAAAAUUAUGCUG------------GUA-----AUUUCGUUACAGUUCACGCAUAAUUC--GUUUAAC---------- ---..................(((((..((((((.((.(((((((((((....------------)))-----)))...))))).))))))))...)))--)).....---------- ( -17.60, z-score = -1.91, R) >droMoj3.scaffold_6500 10770776 86 - 32352404 ---AUCUUUAAAUCGUAAAAUAUGAAAUUGCGUGCAAGUGUAAAAUUAUGCUC------------GUA-----AUUUCGUUACAGUUCACGCAUUAUUC--GUCUAAC---------- ---..................(((((..((((((.((.(((((((((((....------------)))-----)))...))))).))))))))...)))--)).....---------- ( -18.50, z-score = -2.75, R) >droVir3.scaffold_12723 4799120 86 - 5802038 ---AUCUUUAAAUCGUAAAAUAUGAAAUUGCGUGCAAGUGUAAAAUUAUGCUC------------GUA-----AUUUCGUUACAGUUCACGCAUUAUUC--GUCUAAC---------- ---..................(((((..((((((.((.(((((((((((....------------)))-----)))...))))).))))))))...)))--)).....---------- ( -18.50, z-score = -2.75, R) >consensus ___GUCUUCAAAUCGUAAAAUAUGAAAUUGCGUGCAAGUGUUCAAUUAUGUUC_________C__GUAU____AUUUCGCUACACUUCACGCAUUUAAUGCGGUCAGC__________ ............(((((...)))))...((((((.((.(((........................................))).))))))))......................... ( -8.37 = -8.12 + -0.25)


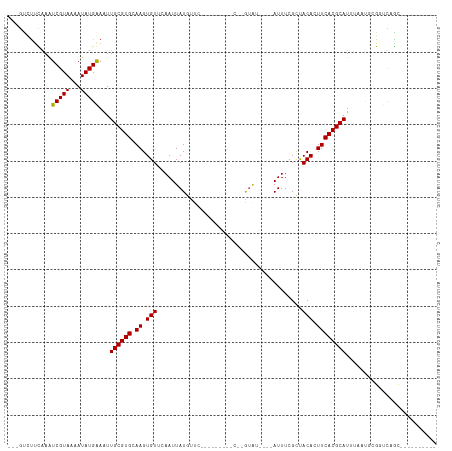
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:00 2011