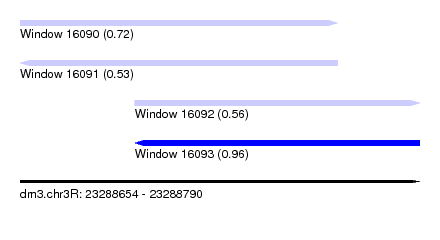
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,288,654 – 23,288,790 |
| Length | 136 |
| Max. P | 0.962474 |

| Location | 23,288,654 – 23,288,762 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 73.41 |
| Shannon entropy | 0.52119 |
| G+C content | 0.47645 |
| Mean single sequence MFE | -32.91 |
| Consensus MFE | -15.90 |
| Energy contribution | -16.75 |
| Covariance contribution | 0.85 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.715566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23288654 108 + 27905053 AUUUGGCAUUUUAUUCGCUUCCUGGUGAUGGUGAUGAUAAUGGUGCUGCUGCU---CCAAGUGCCGGUCAG-UGUGGCAACAAAUGUUGCCAAAGGCUGUGCCACAAUUCGU ....((((((....(((((..(....)..)))))......(((.((....)).---)))))))))((((((-(.(((((((....)))))))...)))).)))......... ( -37.40, z-score = -1.68, R) >droSim1.chr3R 23025278 107 + 27517382 AUUUGGCAUUUUAUUCGCUUCCUGGUAAUGGUGAUGAUGAUGCUGCUGCUGCUGCUCCAAGUGCCGGUCAG-UGUGGCA--CAAUGUUGCCAAAGGCUGUGC-ACA-UUCGU ....((((((....(((((..........)))))....))))))(..((....))..)..(((((((((..-..(((((--......)))))..))))).))-)).-..... ( -31.70, z-score = 0.63, R) >droYak2.chr3R 5628965 94 - 28832112 AUUUGGCAUUUUAUUCGCAUCCUGGUGAUGAUGAUGCU------------------CCAAGUGCCGGUCAUCUGUGGCAACAAAUGUUGCCAAAUCCAGUGCCACAAUUCGU ....(((((((.....(((((...........))))).------------------..)))))))((.((.((((((((((....)))))))....)))))))......... ( -28.30, z-score = -1.43, R) >droEre2.scaffold_4820 5727263 104 - 10470090 AUUUGGCAUUUUAUUCGCUGCCCGGUGAUGCC-AUCGUCAUGCUGCUGCUG------CAAGUGCCGGUCAG-UGUGGCAACAAAUGUUGCCAAAUGCUGUGCCACAAUUCGU ...((((((..(((((((((.(((((((((..-..))))..(((((....)------).)))))))).)))-))(((((((....)))))))))))..))))))........ ( -40.50, z-score = -2.31, R) >droAna3.scaffold_13340 4309180 107 - 23697760 AUUUGGCAUUUUAUUCGUUUCCUGACC-CGGCACGGCUGAUGAUGCUGAUACUUUCCCAA---CUGGUCAG-UGUGGCAACAAAUGUUGCCAAAUGCCGAGCCACAAUUCGU ...((((.........(((....))).-(((((((((.......))))(((((..((...---..))..))-)))((((((....))))))...))))).))))........ ( -33.10, z-score = -1.91, R) >droWil1.scaffold_181089 3531162 89 + 12369635 AUUUGGCAUUUCAUUCGUUUCCUG------------CUGCAGUUGUUGCUACAU---------CUGGUCAA-UGUGGCAACAAAUGUUGCCAG-UUCUGUGCCACAAUUCGU ...((((((............(((------------(.(((.((((((((((((---------.......)-))))))))))).))).).)))-....))))))........ ( -26.49, z-score = -2.17, R) >consensus AUUUGGCAUUUUAUUCGCUUCCUGGUGAUGGUGAUGCUGAUGCUGCUGCUGCU___CCAAGUGCCGGUCAG_UGUGGCAACAAAUGUUGCCAAAUGCUGUGCCACAAUUCGU ...((((((............(((((....................................))))).......(((((((....)))))))......))))))........ (-15.90 = -16.75 + 0.85)

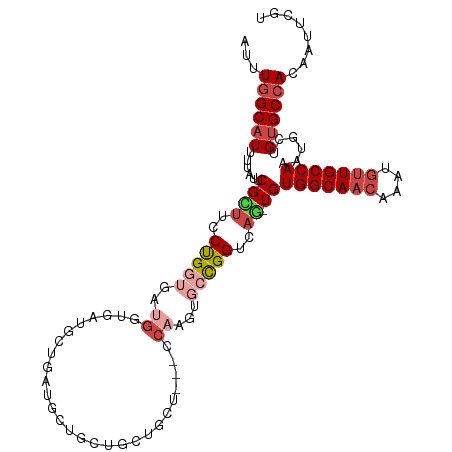
| Location | 23,288,654 – 23,288,762 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 73.41 |
| Shannon entropy | 0.52119 |
| G+C content | 0.47645 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -14.94 |
| Energy contribution | -15.72 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.528041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23288654 108 - 27905053 ACGAAUUGUGGCACAGCCUUUGGCAACAUUUGUUGCCACA-CUGACCGGCACUUGG---AGCAGCAGCACCAUUAUCAUCACCAUCACCAGGAAGCGAAUAAAAUGCCAAAU ........((((((((....(((((((....)))))))..-)))....((...(((---.((....)).))).........((.......))..))........)))))... ( -26.90, z-score = -0.62, R) >droSim1.chr3R 23025278 107 - 27517382 ACGAA-UGU-GCACAGCCUUUGGCAACAUUG--UGCCACA-CUGACCGGCACUUGGAGCAGCAGCAGCAGCAUCAUCAUCACCAUUACCAGGAAGCGAAUAAAAUGCCAAAU .....-(((-((((((....((....)))))--)).))))-......((((..((..((.((....)).))..))......((.......))............)))).... ( -21.60, z-score = 1.56, R) >droYak2.chr3R 5628965 94 + 28832112 ACGAAUUGUGGCACUGGAUUUGGCAACAUUUGUUGCCACAGAUGACCGGCACUUGG------------------AGCAUCAUCAUCACCAGGAUGCGAAUAAAAUGCCAAAU ........(((((((((...(((((((....)))))))..(((((...((......------------------.)).)))))....)))).((....))....)))))... ( -27.90, z-score = -1.63, R) >droEre2.scaffold_4820 5727263 104 + 10470090 ACGAAUUGUGGCACAGCAUUUGGCAACAUUUGUUGCCACA-CUGACCGGCACUUG------CAGCAGCAGCAUGACGAU-GGCAUCACCGGGCAGCGAAUAAAAUGCCAAAU ........(((((.......(((((((....)))))))(.-(((.((((....((------(.(((......)).)...-.)))...)))).))).).......)))))... ( -31.40, z-score = -0.51, R) >droAna3.scaffold_13340 4309180 107 + 23697760 ACGAAUUGUGGCUCGGCAUUUGGCAACAUUUGUUGCCACA-CUGACCAG---UUGGGAAAGUAUCAGCAUCAUCAGCCGUGCCG-GGUCAGGAAACGAAUAAAAUGCCAAAU .((..((.(((((((((((.(((((((....)))))))..-((((...(---((((.......)))))....))))..))))))-))))).))..))............... ( -37.40, z-score = -2.69, R) >droWil1.scaffold_181089 3531162 89 - 12369635 ACGAAUUGUGGCACAGAA-CUGGCAACAUUUGUUGCCACA-UUGACCAG---------AUGUAGCAACAACUGCAG------------CAGGAAACGAAUGAAAUGCCAAAU ........((((((((..-.((....)).(((((((.(((-((.....)---------)))).))))))))))((.------------..(....)...))...)))))... ( -26.10, z-score = -2.71, R) >consensus ACGAAUUGUGGCACAGCAUUUGGCAACAUUUGUUGCCACA_CUGACCGGCACUUGG___AGCAGCAGCAGCAUCAGCAUCACCAUCACCAGGAAGCGAAUAAAAUGCCAAAU ........((((((((....(((((((....)))))))...)))..............................................(....)........)))))... (-14.94 = -15.72 + 0.78)
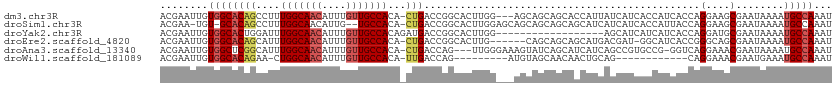
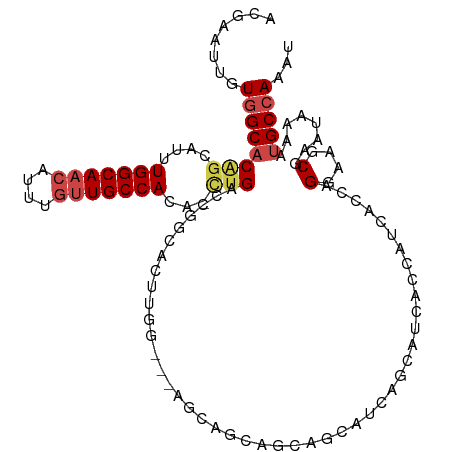
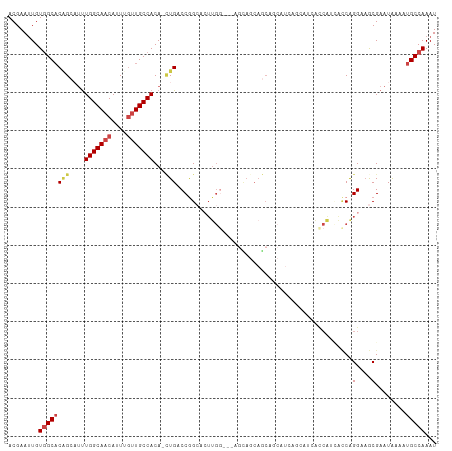
| Location | 23,288,693 – 23,288,790 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 69.99 |
| Shannon entropy | 0.54490 |
| G+C content | 0.50633 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -14.16 |
| Energy contribution | -15.38 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.563398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23288693 97 + 27905053 ---AUGGUGCUGCUGCUCCAAGUGCCGGUCAG-UGUGGCAACAAAUGUUGCCAAAGGCUGUGCCACAAUUCGUUUCAUUUGCGGCCAACGGGAACGGGAAC------ ---.(((.((....)).))).(((((((((..-..(((((((....)))))))..))))).).)))..((((((((..(((....)))..))))))))...------ ( -33.80, z-score = -0.38, R) >droSim1.chr3R 23025317 93 + 27517382 AUGCUGCUGCUGCUGCUCCAAGUGCCGGUCAG-UGUGGCAC--AAUGUUGCCAAAGGCUGUGC-ACA-UUCGUUUCAUUUGCGGCC-ACGGAACGGAAC-------- ..((.((....)).)).....(((((((((..-..(((((.--.....)))))..))))).))-)).-((((((((..........-..))))))))..-------- ( -30.20, z-score = 0.65, R) >droYak2.chr3R 5628992 88 - 28832112 ------AUGAUGAUGCUCCAAGUGCCGGUCAUCUGUGGCAACAAAUGUUGCCAAAUCCAGUGCCACAAUUCGUUUCAUUUGCGGCAAACGGCAG------------- ------................(((((.....((((((((((....)))))))....)))((((.(((..........))).))))..))))).------------- ( -27.20, z-score = -1.33, R) >droEre2.scaffold_4820 5727301 84 - 10470090 ------AUGCUGCUGCUGCAAGUGCCGGUCAG-UGUGGCAACAAAUGUUGCCAAAUGCUGUGCCACAAUUCGUUUCAUUUGCGGCCCACGG---------------- ------...(((..((((((((((..((((((-(.(((((((....)))))))...)))).)))((.....))..))))))))))...)))---------------- ( -32.10, z-score = -1.51, R) >droAna3.scaffold_13340 4309218 85 - 23697760 ---AUGAUGCUGAUACUUUCCCAACUGGUCAG-UGUGGCAACAAAUGUUGCCAAAUGCCGAGCCACAAUUCGUUUAAUUUGCGGCCACC------------------ ---...((((((((.............)))))-)))((((((....)))))).......(.(((.(((..........))).))))...------------------ ( -22.02, z-score = -0.87, R) >droWil1.scaffold_181089 3531189 96 + 12369635 ---------CAGUUGUUGCUACAUCUGGUCAA-UGUGGCAACAAAUGUUGCCAG-UUCUGUGCCACAAUUCGUUUAAUUUGUGGCUAGAGCACAUUGAGAAGUGGAC ---------(((.(((....))).))).((((-(((((((((....)))))).(-(((((.(((((((..........))))))))))))))))))))......... ( -38.40, z-score = -4.29, R) >consensus ______AUGCUGCUGCUCCAAGUGCCGGUCAG_UGUGGCAACAAAUGUUGCCAAAUGCUGUGCCACAAUUCGUUUCAUUUGCGGCCAACGG_A______________ ........................(((........(((((((....)))))))........(((.(((..........))).)))...)))................ (-14.16 = -15.38 + 1.22)

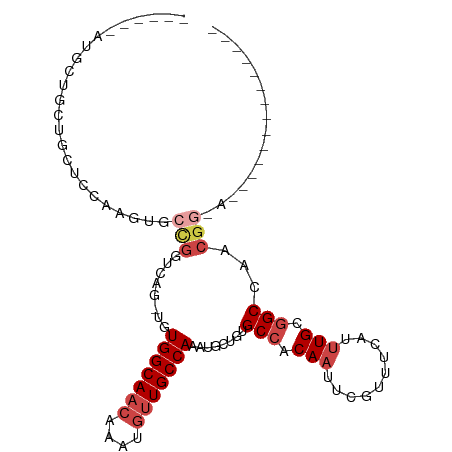
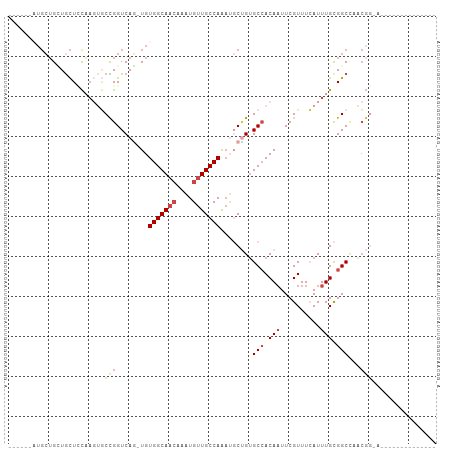
| Location | 23,288,693 – 23,288,790 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 69.99 |
| Shannon entropy | 0.54490 |
| G+C content | 0.50633 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -18.53 |
| Energy contribution | -19.40 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23288693 97 - 27905053 ------GUUCCCGUUCCCGUUGGCCGCAAAUGAAACGAAUUGUGGCACAGCCUUUGGCAACAUUUGUUGCCACA-CUGACCGGCACUUGGAGCAGCAGCACCAU--- ------(((((.((.((.(((((((((((..........))))))).))))...(((((((....)))))))..-......)).))..)))))...........--- ( -32.80, z-score = -1.40, R) >droSim1.chr3R 23025317 93 - 27517382 --------GUUCCGUUCCGU-GGCCGCAAAUGAAACGAA-UGU-GCACAGCCUUUGGCAACAUU--GUGCCACA-CUGACCGGCACUUGGAGCAGCAGCAGCAGCAU --------(((((((((((.-(((.(((.((.......)-).)-))...)))..))).)))...--(((((...-......)))))..))))).((....))..... ( -26.90, z-score = 0.89, R) >droYak2.chr3R 5628992 88 + 28832112 -------------CUGCCGUUUGCCGCAAAUGAAACGAAUUGUGGCACUGGAUUUGGCAACAUUUGUUGCCACAGAUGACCGGCACUUGGAGCAUCAUCAU------ -------------.(((((..((((((((..........))))))))(((....(((((((....)))))))))).....)))))................------ ( -29.20, z-score = -1.89, R) >droEre2.scaffold_4820 5727301 84 + 10470090 ----------------CCGUGGGCCGCAAAUGAAACGAAUUGUGGCACAGCAUUUGGCAACAUUUGUUGCCACA-CUGACCGGCACUUGCAGCAGCAGCAU------ ----------------..(((((((((((..........))))))).(((((..((....))..))))))))).-(((....((....)).....)))...------ ( -28.60, z-score = -0.77, R) >droAna3.scaffold_13340 4309218 85 + 23697760 ------------------GGUGGCCGCAAAUUAAACGAAUUGUGGCUCGGCAUUUGGCAACAUUUGUUGCCACA-CUGACCAGUUGGGAAAGUAUCAGCAUCAU--- ------------------(((((((((((.((....)).))))))))(((....(((((((....)))))))..-)))))).(((((.......))))).....--- ( -27.80, z-score = -1.90, R) >droWil1.scaffold_181089 3531189 96 - 12369635 GUCCACUUCUCAAUGUGCUCUAGCCACAAAUUAAACGAAUUGUGGCACAGAA-CUGGCAACAUUUGUUGCCACA-UUGACCAGAUGUAGCAACAACUG--------- .........(((((((......(((((((.((....)).)))))))......-..((((((....)))))))))-)))).(((.(((....))).)))--------- ( -29.40, z-score = -3.27, R) >consensus ______________U_CCGUUGGCCGCAAAUGAAACGAAUUGUGGCACAGCAUUUGGCAACAUUUGUUGCCACA_CUGACCGGCACUUGGAGCAGCAGCAU______ ..................(((((((((((..........))))))).(((....(((((((....)))))))...)))..))))....................... (-18.53 = -19.40 + 0.87)

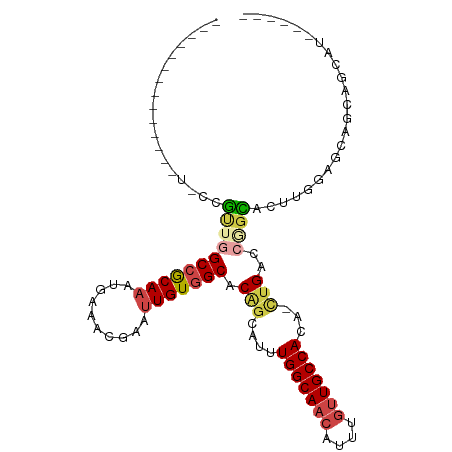
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:48:09 2011