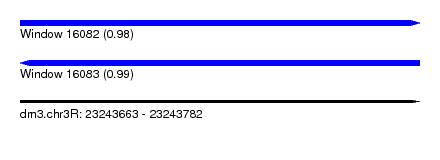
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,243,663 – 23,243,782 |
| Length | 119 |
| Max. P | 0.994641 |

| Location | 23,243,663 – 23,243,782 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.65 |
| Shannon entropy | 0.09712 |
| G+C content | 0.43196 |
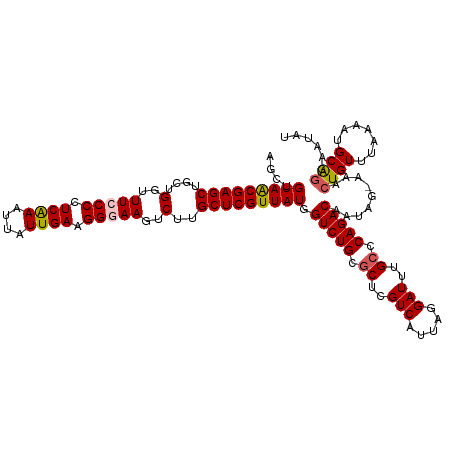
| Mean single sequence MFE | -32.70 |
| Consensus MFE | -29.96 |
| Energy contribution | -30.96 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23243663 119 + 27905053 AUAUUCCUGCAUUUUAAACAGUUU-CUAUUGUCUGGGCAAAUCCUAAUGACGAGCGCAGACCAUAACGAGCAAGACUUCCCUUCAAAAAUUUGAGGGGAAACCAGCAGCUCGUUACAGCU ........((..............-.....(((((.((...((.....))...)).)))))..((((((((.....((((((((((....)))))))))).......))))))))..)). ( -34.80, z-score = -3.11, R) >droSim1.chr3R 22977108 119 + 27517382 AUAUUCCUGCAUUUUAAACAGUUU-CUAUUGUCUGGGCAAAUCCUAAUGACGAGCGCAGACCAUAACGAGCAAGACUUCCCUUCAAUAAUUUGAGGGGAAAGCAGCAGCUCGUUACAGCU ........((..............-.....(((((.((...((.....))...)).)))))..((((((((..(..((((((((((....))))))))))..)....))))))))..)). ( -36.40, z-score = -3.25, R) >droSec1.super_13 1987354 119 + 2104621 AUAUUCCUGCAUUUUAAACAGUUU-CUAUUGUCUGGGCAAAUCCUAAUGACGAGCGCAGACCAUAACGAGCAAGACUUCCCUUCAAUAAUUUGAGGGGAAACCAGCAGCUCGCUACAGCU ......((((..........((((-.(((.(((((.((...((.....))...)).))))).)))..)))).....((((((((((....))))))))))....))))...((....)). ( -33.20, z-score = -2.48, R) >droYak2.chr3R 5580917 120 - 28832112 AUAGUCGUGCAUUUUAAACAGUUUUCUAUUGUCUGGGCAAAUCCUAAUGACGAGCGCAGACCAUAACGAGCAAGAGUUCCCUUCAAUAAUUUGAGGGUAAACCAGCAGCUCGUUACUGCU ...(((((((.........((....)).(((((((((.....))))..))))))))).)))..((((((((....(((.(((((((....)))))))..))).....))))))))..... ( -32.00, z-score = -1.76, R) >droEre2.scaffold_4820 5681684 118 - 10470090 AUAUUCCCGCACUUUAAACAGUUUUCUAUUGUCUGG-CAAAUCCUAAUGACGAGUGCAGACCAUAACGAGCAAGACUUCCCUUCAAUAAUUCGAGGG-AAACCAGCAGCUCGUUACCGCU ........((((((.....((....))...(((.((-.....))....)))))))))......((((((((.....(((((((.........)))))-)).......))))))))..... ( -27.10, z-score = -1.70, R) >consensus AUAUUCCUGCAUUUUAAACAGUUU_CUAUUGUCUGGGCAAAUCCUAAUGACGAGCGCAGACCAUAACGAGCAAGACUUCCCUUCAAUAAUUUGAGGGGAAACCAGCAGCUCGUUACAGCU ........((....................(((((.((...((.....))...)).)))))..((((((((.....((((((((((....)))))))))).......))))))))..)). (-29.96 = -30.96 + 1.00)

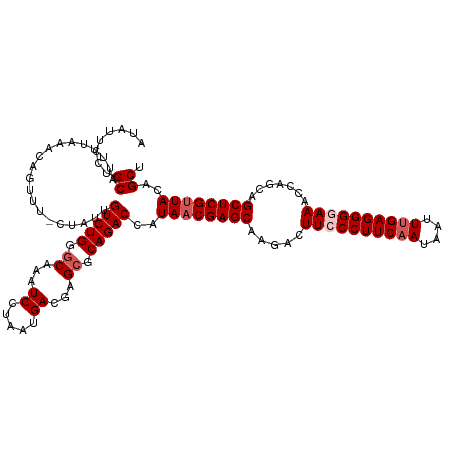
| Location | 23,243,663 – 23,243,782 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.65 |
| Shannon entropy | 0.09712 |
| G+C content | 0.43196 |
| Mean single sequence MFE | -39.68 |
| Consensus MFE | -35.52 |
| Energy contribution | -35.84 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.994641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23243663 119 - 27905053 AGCUGUAACGAGCUGCUGGUUUCCCCUCAAAUUUUUGAAGGGAAGUCUUGCUCGUUAUGGUCUGCGCUCGUCAUUAGGAUUUGCCCAGACAAUAG-AAACUGUUUAAAAUGCAGGAAUAU ....(((((((((....(..(((((.((((....)))).)))))..)..))))))))).(((((.((..(((.....)))..)).))))).....-...((((.......))))...... ( -41.10, z-score = -3.60, R) >droSim1.chr3R 22977108 119 - 27517382 AGCUGUAACGAGCUGCUGCUUUCCCCUCAAAUUAUUGAAGGGAAGUCUUGCUCGUUAUGGUCUGCGCUCGUCAUUAGGAUUUGCCCAGACAAUAG-AAACUGUUUAAAAUGCAGGAAUAU ....(((((((((....((((.(((.((((....)))).)))))))...))))))))).(((((.((..(((.....)))..)).))))).....-...((((.......))))...... ( -38.70, z-score = -3.08, R) >droSec1.super_13 1987354 119 - 2104621 AGCUGUAGCGAGCUGCUGGUUUCCCCUCAAAUUAUUGAAGGGAAGUCUUGCUCGUUAUGGUCUGCGCUCGUCAUUAGGAUUUGCCCAGACAAUAG-AAACUGUUUAAAAUGCAGGAAUAU ....(((((((((....(..(((((.((((....)))).)))))..)..))))))))).(((((.((..(((.....)))..)).))))).....-...((((.......))))...... ( -41.10, z-score = -3.25, R) >droYak2.chr3R 5580917 120 + 28832112 AGCAGUAACGAGCUGCUGGUUUACCCUCAAAUUAUUGAAGGGAACUCUUGCUCGUUAUGGUCUGCGCUCGUCAUUAGGAUUUGCCCAGACAAUAGAAAACUGUUUAAAAUGCACGACUAU .((((((((((((....((((..(((((((....))).))))))))...))))))))).(((((.((..(((.....)))..)).)))))(((((....))))).....)))........ ( -36.40, z-score = -2.79, R) >droEre2.scaffold_4820 5681684 118 + 10470090 AGCGGUAACGAGCUGCUGGUUU-CCCUCGAAUUAUUGAAGGGAAGUCUUGCUCGUUAUGGUCUGCACUCGUCAUUAGGAUUUG-CCAGACAAUAGAAAACUGUUUAAAGUGCGGGAAUAU .((.(((((((((....(..((-(((((((....))).))))))..)..))))))))).))(((((((.(((....((.....-)).)))(((((....)))))...)))))))...... ( -41.10, z-score = -3.39, R) >consensus AGCUGUAACGAGCUGCUGGUUUCCCCUCAAAUUAUUGAAGGGAAGUCUUGCUCGUUAUGGUCUGCGCUCGUCAUUAGGAUUUGCCCAGACAAUAG_AAACUGUUUAAAAUGCAGGAAUAU ....(((((((((....(..(((((.((((....)))).)))))..)..))))))))).(((((.((..(((.....)))..)).))))).........((((.......))))...... (-35.52 = -35.84 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:48:01 2011