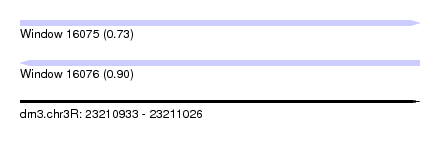
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,210,933 – 23,211,026 |
| Length | 93 |
| Max. P | 0.896131 |

| Location | 23,210,933 – 23,211,026 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.45 |
| Shannon entropy | 0.44633 |
| G+C content | 0.54109 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -23.26 |
| Energy contribution | -23.11 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.15 |
| Mean z-score | -0.68 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.733637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23210933 93 + 27905053 UGGCAUCCAAGCGUUUCCGCUCCAUUUGCGGGCCGUCAAAGCAACGGCCUCAGAAUUGAAACAUUGCCAGGACC----AGCUCCUUACCUUCAGGUU------ (((((....((((....))))...((((.(((((((.......)))))))))))..........)))))(((..----...)))..(((....))).------ ( -26.50, z-score = -0.70, R) >droSim1.chr3R 22944435 97 + 27517382 UGGCAUCCAAGCGUUUCCGCUCCAUUUGCGGGCCGUCAAAGCAACGGCCUCGGAAUUGAAACAUUGCCAGGACCUCAGGACUCCAUACCUUCAGGUU------ (((((......((.((((((.......))(((((((.......))))))).)))).))......))))).(((((.(((........)))..)))))------ ( -32.00, z-score = -1.62, R) >droSec1.super_13 1954543 89 + 2104621 UGGCAUCCAAGCGUUUCCGCUCCAUUUGCGGGCCGUCAAAGCAACGGCCUCCGAAUUGAAACAUUGCCAGGAC--------UCCAUACCUUCAGGUU------ (((((.....(((....)))((.(((((.(((((((.......))))))).))))).)).....)))))....--------.....(((....))).------ ( -27.20, z-score = -1.44, R) >droYak2.chr3R 5545266 95 - 28832112 UGGCAUCCAAGCGUUUCCGCUCCAUUUGCGGGCCGUCAAAGCAACGGCCUCGGAAUUGAAACAUUGCCAGGACC--AGCUUUCCUUUCCUUCUGGUU------ (((((......((.((((((.......))(((((((.......))))))).)))).))......))))).((((--((.............))))))------ ( -30.32, z-score = -1.22, R) >droEre2.scaffold_4820 5644007 93 - 10470090 UGGCAUCCAAGCGUUUCCGCUCCAUUUGCGGGCCGUCAAAGCAACGGCCUCGGAAUUGAAACAUUGCCAGGACC--AGCU--ACUUUCCUUCAGGUU------ (((((......((.((((((.......))(((((((.......))))))).)))).))......))))).((((--....--...........))))------ ( -26.96, z-score = -0.43, R) >droAna3.scaffold_13340 4230006 80 - 23697760 UGGCAGGGCAGCAUUUCCGCUCCAUUUGCGGGCCGUCAAAGGAAAGGCCUCGGAAUUGAAACAUUGCCAACCU----UCGAGCU------------------- ((((((.....((.((((((.......))(((((.((....))..))))).)))).)).....))))))....----.......------------------- ( -24.90, z-score = 0.01, R) >dp4.chr2 8868696 87 + 30794189 UGGCAGAGCAACAUUUCCGCUCCAUUUGCGGGCCGUCAAAGCAACGGCUCCGGAAUUGAAACAUUGCCAACAUC--AUGCAGCCAGGCC-------------- (((((((((.........))))((.((.((((((((.......))))).))).)).))......))))).....--..((......)).-------------- ( -26.30, z-score = -0.67, R) >droPer1.super_0 2681492 87 + 11822988 UGGCAGAGAAACAUUUCCGCUCCAUUUGCGGGCCGUCAAAGCAACGGCUCCGGAAUUGAAACAUUGCCAACAUC--AUGCAGCCAGGCC-------------- ((((((.(...((.((((((.......))(((((((.......))))).)))))).))...).)))))).....--..((......)).-------------- ( -25.70, z-score = -0.75, R) >droWil1.scaffold_181089 3445047 100 + 12369635 UGGCAG-GCAGCAUUUCCGCUCCAUUUGCGGGCAGUCAAAGCAACGGCCUCGGAAUUGAAACAUUGCCAACAUC--AUGCAGCAGCAUCAGCAGCCUCUCAUU .(((.(-((((((.((((((.......))((((.((.......)).)))).)))).)).....)))))......--((((....)))).....)))....... ( -25.60, z-score = 0.71, R) >consensus UGGCAUCCAAGCGUUUCCGCUCCAUUUGCGGGCCGUCAAAGCAACGGCCUCGGAAUUGAAACAUUGCCAGGACC__AUGCAGCCAUACCUUCAGGUU______ (((((......((.((((((.......))(((((((.......))))))).)))).))......))))).................................. (-23.26 = -23.11 + -0.15)


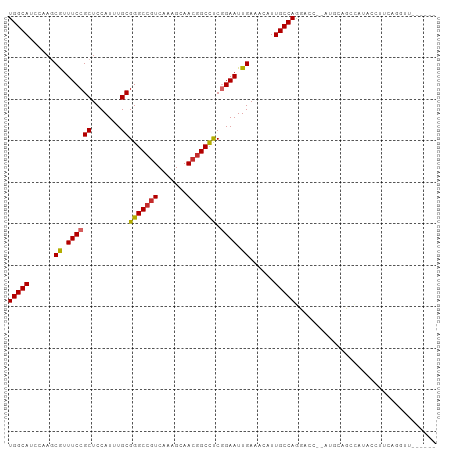
| Location | 23,210,933 – 23,211,026 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.45 |
| Shannon entropy | 0.44633 |
| G+C content | 0.54109 |
| Mean single sequence MFE | -33.24 |
| Consensus MFE | -23.63 |
| Energy contribution | -23.68 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
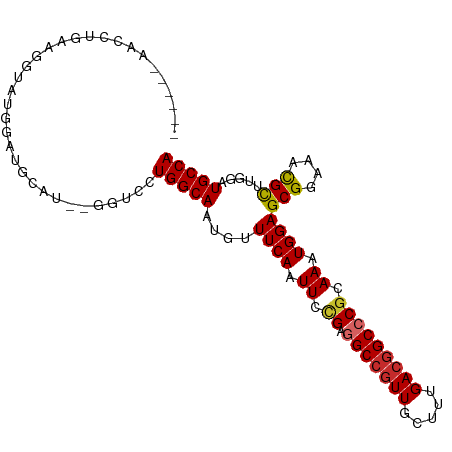
>dm3.chr3R 23210933 93 - 27905053 ------AACCUGAAGGUAAGGAGCU----GGUCCUGGCAAUGUUUCAAUUCUGAGGCCGUUGCUUUGACGGCCCGCAAAUGGAGCGGAAACGCUUGGAUGCCA ------........((((.((.(((----(.((..((((((((((((....))))).)))))))..)))))))).....(.(((((....))))).).)))). ( -36.40, z-score = -2.17, R) >droSim1.chr3R 22944435 97 - 27517382 ------AACCUGAAGGUAUGGAGUCCUGAGGUCCUGGCAAUGUUUCAAUUCCGAGGCCGUUGCUUUGACGGCCCGCAAAUGGAGCGGAAACGCUUGGAUGCCA ------.((((..(((........))).))))...((((......((.((.((.(((((((.....))))))))).)).))(((((....)))))...)))). ( -37.10, z-score = -1.87, R) >droSec1.super_13 1954543 89 - 2104621 ------AACCUGAAGGUAUGGA--------GUCCUGGCAAUGUUUCAAUUCGGAGGCCGUUGCUUUGACGGCCCGCAAAUGGAGCGGAAACGCUUGGAUGCCA ------.(((....))).(((.--------((((.(((...((((((.(((((..((((((.....))))))))).)).))))))(....)))).)))).))) ( -35.00, z-score = -2.25, R) >droYak2.chr3R 5545266 95 + 28832112 ------AACCAGAAGGAAAGGAAAGCU--GGUCCUGGCAAUGUUUCAAUUCCGAGGCCGUUGCUUUGACGGCCCGCAAAUGGAGCGGAAACGCUUGGAUGCCA ------.......((((..((....))--..))))((((......((.((.((.(((((((.....))))))))).)).))(((((....)))))...)))). ( -36.20, z-score = -1.93, R) >droEre2.scaffold_4820 5644007 93 + 10470090 ------AACCUGAAGGAAAGU--AGCU--GGUCCUGGCAAUGUUUCAAUUCCGAGGCCGUUGCUUUGACGGCCCGCAAAUGGAGCGGAAACGCUUGGAUGCCA ------..((....)).....--.((.--.((((.(((...((((((.((.((.(((((((.....))))))))).)).))))))(....)))).)))))).. ( -33.50, z-score = -1.43, R) >droAna3.scaffold_13340 4230006 80 + 23697760 -------------------AGCUCGA----AGGUUGGCAAUGUUUCAAUUCCGAGGCCUUUCCUUUGACGGCCCGCAAAUGGAGCGGAAAUGCUGCCCUGCCA -------------------.......----.(((.((((..(((((..(((((.((((((......)).))))......)))))..)))))..))))..))). ( -25.40, z-score = -0.16, R) >dp4.chr2 8868696 87 - 30794189 --------------GGCCUGGCUGCAU--GAUGUUGGCAAUGUUUCAAUUCCGGAGCCGUUGCUUUGACGGCCCGCAAAUGGAGCGGAAAUGUUGCUCUGCCA --------------(((..(((.((((--..((....)).(((((((.((.(((.((((((.....))))))))).)).)))))))...)))).)))..))). ( -32.90, z-score = -1.23, R) >droPer1.super_0 2681492 87 - 11822988 --------------GGCCUGGCUGCAU--GAUGUUGGCAAUGUUUCAAUUCCGGAGCCGUUGCUUUGACGGCCCGCAAAUGGAGCGGAAAUGUUUCUCUGCCA --------------(((..((..((((--..((....)).(((((((.((.(((.((((((.....))))))))).)).)))))))...))))..))..))). ( -29.50, z-score = -0.54, R) >droWil1.scaffold_181089 3445047 100 - 12369635 AAUGAGAGGCUGCUGAUGCUGCUGCAU--GAUGUUGGCAAUGUUUCAAUUCCGAGGCCGUUGCUUUGACUGCCCGCAAAUGGAGCGGAAAUGCUGC-CUGCCA ..((..((((.((.....(((((.(((--..(((.((((..(((((......))))).(((.....))))))).))).))).)))))....)).))-))..)) ( -33.20, z-score = -0.64, R) >consensus ______AACCUGAAGGUAUGGAUGCAU__GGUCCUGGCAAUGUUUCAAUUCCGAGGCCGUUGCUUUGACGGCCCGCAAAUGGAGCGGAAACGCUUGGAUGCCA ..................................(((((....((((.((.((.(((((((.....))))))))).)).))))(((....))).....))))) (-23.63 = -23.68 + 0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:47:55 2011