| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,203,157 – 23,203,261 |
| Length | 104 |
| Max. P | 0.905736 |

| Location | 23,203,157 – 23,203,261 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 86.04 |
| Shannon entropy | 0.27521 |
| G+C content | 0.43484 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -22.45 |
| Energy contribution | -22.21 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
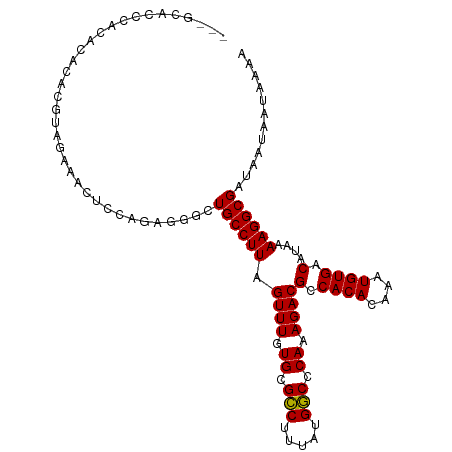
>dm3.chr3R 23203157 104 + 27905053 UUUUAUUAUUAUCGCCUUUUAUGUCACAUUUGUGUGGCGUCUUUGGGCCAUAAAGGCGCACAAACUAAGGCAGCCCUCUGGAGUUUCAAUGUGUGUGUAAGUGC--- ........((((.((((...((((((((....))))))))....)))).))))(.(((((((((((.(((....)))....))))....))))))).)......--- ( -27.60, z-score = -0.72, R) >droSim1.chr3R 22936703 104 + 27517382 UUUUAUUAUUAUCGCCUUUUAUGUCACAUUUGUGUGGCGUCUUUGGGCCAUAAAGGCGCACAAACUAAGGCAGCCCUCUGGUGUUUCUAUGUGUGUGUAAGUGC--- ........((((.((((...((((((((....))))))))....)))).))))..(((((((.....(((((.((....))))))).....)))))))......--- ( -29.60, z-score = -1.47, R) >droSec1.super_13 1946853 104 + 2104621 UUUUAUUAUUAUCGCCUUUUAUGUCACAUUUGUGUGGCGUCUUUGGGCCAUAAAGGCGCACAAACUAAGGCAGCCCUCUGGUGUUUCUAUGUGUGUGUAAGUGC--- ........((((.((((...((((((((....))))))))....)))).))))..(((((((.....(((((.((....))))))).....)))))))......--- ( -29.60, z-score = -1.47, R) >droYak2.chr3R 5536369 99 - 28832112 UUUUAUUAUUAUCGCCUUUUAUGUCACAUUUGUGUGGCGUCUUUGGGCCAUAAAGGCGCACAAACUAAGGCAGCUCUCUGGAGUUUCUACGUGUGUGUG-------- ........((((.((((...((((((((....))))))))....)))).))))..(((((((.....(((.(((((....))))))))...))))))).-------- ( -31.10, z-score = -2.89, R) >droEre2.scaffold_4820 5636339 104 - 10470090 UUUUAUUAUUAUCGCCUUUUAUGUCACAUUUGUGUGGCGUCUUUGGGCCAUAAAGGCGCACAAACUAAGGCAGCUCUCUGGAGCUUCUACGUGUGCGUGUGUGC--- ........((((.((((...((((((((....))))))))....)))).))))(.(((((((.....(((.(((((....))))))))...))))))).)....--- ( -35.70, z-score = -2.91, R) >droAna3.scaffold_13340 4220710 104 - 23697760 UUUUAUUAUUAUCGCCUUUUAUGUCACAUUUGUGUGGCGUCUUUGAGUCAUAAAGACGCACAAACUAAGGCAACUCUAUGCCCUCUAUGCGUGGGUGUGGCUAU--- .............(((((...(((((((....))))(((((((((.....))))))))))))....)))))....((((((((.(.....).))))))))....--- ( -31.80, z-score = -3.01, R) >droPer1.super_0 2673145 101 + 11822988 UUUUAUUAUUAUCGCCUUUUAUGUCACAUUUGUGUGGCGUCUUUGAGUCAUAAAGACGCACAAACUAAGGCAGCCCUA------CUAUGCGUGUGUGUGGCCGGUGU .........(((((((((...(((((((....))))(((((((((.....))))))))))))....))))).(((.((------(.........))).))).)))). ( -28.90, z-score = -1.35, R) >dp4.chr2 8860371 101 + 30794189 UUUUAUUAUUAUCGCCUUUUAUGUCACAUUUGUGUGGCGUCUUUGAGUCAUAAAGACGCACAAACUAAGGCAGCCGUA------CUAUGCGUGUGUGUGGCCGGUGU .............(((((...(((((((....))))(((((((((.....))))))))))))....))))).((((..------((((((....)))))).)))).. ( -30.30, z-score = -1.56, R) >consensus UUUUAUUAUUAUCGCCUUUUAUGUCACAUUUGUGUGGCGUCUUUGGGCCAUAAAGGCGCACAAACUAAGGCAGCCCUCUGGAGUUUCUACGUGUGUGUGAGUGC___ .............(((((...(((((((....))))(((((((((.....))))))))))))....))))).................................... (-22.45 = -22.21 + -0.23)


| Location | 23,203,157 – 23,203,261 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 86.04 |
| Shannon entropy | 0.27521 |
| G+C content | 0.43484 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -18.78 |
| Energy contribution | -18.31 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23203157 104 - 27905053 ---GCACUUACACACACAUUGAAACUCCAGAGGGCUGCCUUAGUUUGUGCGCCUUUAUGGCCCAAAGACGCCACACAAAUGUGACAUAAAAGGCGAUAAUAAUAAAA ---((((..((........((......))((((....)))).))..))))((((((.((((........))))(((....))).....))))))............. ( -25.30, z-score = -1.75, R) >droSim1.chr3R 22936703 104 - 27517382 ---GCACUUACACACACAUAGAAACACCAGAGGGCUGCCUUAGUUUGUGCGCCUUUAUGGCCCAAAGACGCCACACAAAUGUGACAUAAAAGGCGAUAAUAAUAAAA ---((((..((.........(......).((((....)))).))..))))((((((.((((........))))(((....))).....))))))............. ( -24.60, z-score = -1.85, R) >droSec1.super_13 1946853 104 - 2104621 ---GCACUUACACACACAUAGAAACACCAGAGGGCUGCCUUAGUUUGUGCGCCUUUAUGGCCCAAAGACGCCACACAAAUGUGACAUAAAAGGCGAUAAUAAUAAAA ---((((..((.........(......).((((....)))).))..))))((((((.((((........))))(((....))).....))))))............. ( -24.60, z-score = -1.85, R) >droYak2.chr3R 5536369 99 + 28832112 --------CACACACACGUAGAAACUCCAGAGAGCUGCCUUAGUUUGUGCGCCUUUAUGGCCCAAAGACGCCACACAAAUGUGACAUAAAAGGCGAUAAUAAUAAAA --------((((.((..((((...(((....)))))))....)).))))(((((((.((((........))))(((....))).....)))))))............ ( -23.30, z-score = -2.27, R) >droEre2.scaffold_4820 5636339 104 + 10470090 ---GCACACACGCACACGUAGAAGCUCCAGAGAGCUGCCUUAGUUUGUGCGCCUUUAUGGCCCAAAGACGCCACACAAAUGUGACAUAAAAGGCGAUAAUAAUAAAA ---(((((.((((....))...(((((....)))))......)).)))))((((((.((((........))))(((....))).....))))))............. ( -29.10, z-score = -2.79, R) >droAna3.scaffold_13340 4220710 104 + 23697760 ---AUAGCCACACCCACGCAUAGAGGGCAUAGAGUUGCCUUAGUUUGUGCGUCUUUAUGACUCAAAGACGCCACACAAAUGUGACAUAAAAGGCGAUAAUAAUAAAA ---...(((.......(((((..((((((......)))))).((.((.((((((((.......)))))))))).))..)))))........)))............. ( -28.66, z-score = -3.13, R) >droPer1.super_0 2673145 101 - 11822988 ACACCGGCCACACACACGCAUAG------UAGGGCUGCCUUAGUUUGUGCGUCUUUAUGACUCAAAGACGCCACACAAAUGUGACAUAAAAGGCGAUAAUAAUAAAA .....((((..((.........)------)..))))(((((....(((((((((((.......))))))))((((....)))))))...)))))............. ( -25.70, z-score = -1.75, R) >dp4.chr2 8860371 101 - 30794189 ACACCGGCCACACACACGCAUAG------UACGGCUGCCUUAGUUUGUGCGUCUUUAUGACUCAAAGACGCCACACAAAUGUGACAUAAAAGGCGAUAAUAAUAAAA .....((((..((.........)------)..))))(((((....(((((((((((.......))))))))((((....)))))))...)))))............. ( -24.30, z-score = -1.55, R) >consensus ___GCACCCACACACACGUAGAAACUCCAGAGGGCUGCCUUAGUUUGUGCGCCUUUAUGGCCCAAAGACGCCACACAAAUGUGACAUAAAAGGCGAUAAUAAUAAAA ...................................((((((.((((.((.(((.....))).)).))))(.((((....)))).)....))))))............ (-18.78 = -18.31 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:47:54 2011