
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,803,072 – 8,803,166 |
| Length | 94 |
| Max. P | 0.735478 |

| Location | 8,803,072 – 8,803,166 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 74.09 |
| Shannon entropy | 0.44236 |
| G+C content | 0.56254 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -17.69 |
| Energy contribution | -18.70 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.626414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8803072 94 + 23011544 CUGCCAAAGCCGAGAUGGAUGACUUGAAUGGCCGGGCAG--------AAGGACGGAAGGUUCGCCUGGAUGGAUGGGCGAGUAU-----UUCGGUUCGGCUGGAUGG ...(((.(((((((.((((..(((((..((.((((((.(--------((...(....).))))))))).....))..)))))..-----)))).)))))))...))) ( -34.30, z-score = -2.19, R) >droSim1.chr2L 8581203 94 + 22036055 CUGCCAAAGCCGAGAUGGAUGACUUGAAUGGCCGGGCAG--------AAGGACAGAAGGUUCGCCUGGAUGGAUGGGCGAGUAU-----UUCGGUUCGGCUGGAUGG ...(((.(((((((.((((..(((((..((.((((((..--------..((((.....)))))))))).....))..)))))..-----)))).)))))))...))) ( -32.30, z-score = -1.64, R) >droSec1.super_3 4275023 93 + 7220098 CUGCCAAAGCCGAGAUGGAUGACUUGAAUGGCCGGGCAG--------AAGGACAGAAGGUUCGCCUGGAUGGAUGGGCGAGUAU-----UUCGGUUCGGCUGGAUG- ...(((..((((((.((((..(((((..((.((((((..--------..((((.....)))))))))).....))..)))))..-----)))).)))))))))...- ( -31.00, z-score = -1.33, R) >droEre2.scaffold_4929 9391464 107 + 26641161 CUGCCAAAGCCGAGAUGGAUGACUUGGAUGGCCGGGCAGGAGGACAGAAGGACAGAAGGUUCGCCUGGAUGGAUGGGCGAGUAUAGUAUUUCGGUUCGGCUGGAUGG ..(((..((((((((((....(((((.....(((..((((.((((.............)))).))))..))).....)))))....)))))))))).)))....... ( -34.42, z-score = -1.39, R) >droAna3.scaffold_12916 10465326 76 + 16180835 CUGCCAAAUCCGAGAUGGAUGGCUUGGAUGGACAUUUGGAUGGAUGGAUGGUUUAGGUUUUGGCCUGGCUGGAUGG------------------------------- ...(((..((((((........))))))))).((((..(..((..((((.......))))...))...)..)))).------------------------------- ( -18.10, z-score = -0.09, R) >consensus CUGCCAAAGCCGAGAUGGAUGACUUGAAUGGCCGGGCAG________AAGGACAGAAGGUUCGCCUGGAUGGAUGGGCGAGUAU_____UUCGGUUCGGCUGGAUGG (((((...((((....((....))....))))..)))))....................((((((((......))))))))........(((((.....)))))... (-17.69 = -18.70 + 1.01)

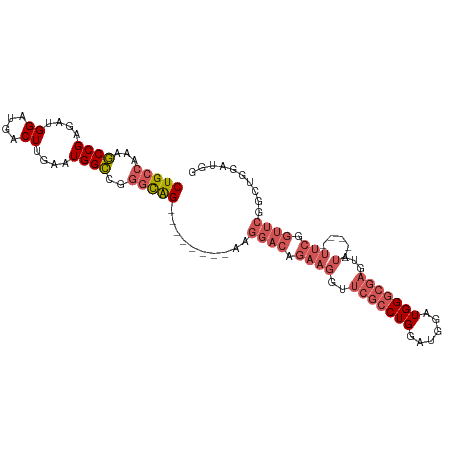

| Location | 8,803,072 – 8,803,166 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 74.09 |
| Shannon entropy | 0.44236 |
| G+C content | 0.56254 |
| Mean single sequence MFE | -20.07 |
| Consensus MFE | -10.88 |
| Energy contribution | -13.88 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8803072 94 - 23011544 CCAUCCAGCCGAACCGAA-----AUACUCGCCCAUCCAUCCAGGCGAACCUUCCGUCCUU--------CUGCCCGGCCAUUCAAGUCAUCCAUCUCGGCUUUGGCAG .......((((((((((.-----....(((((..........))))).....(((..(..--------..)..)))..................)))).)))))).. ( -23.50, z-score = -2.40, R) >droSim1.chr2L 8581203 94 - 22036055 CCAUCCAGCCGAACCGAA-----AUACUCGCCCAUCCAUCCAGGCGAACCUUCUGUCCUU--------CUGCCCGGCCAUUCAAGUCAUCCAUCUCGGCUUUGGCAG .......((((((((((.-----....(((((..........))))).............--------......(((.......))).......)))).)))))).. ( -22.00, z-score = -1.69, R) >droSec1.super_3 4275023 93 - 7220098 -CAUCCAGCCGAACCGAA-----AUACUCGCCCAUCCAUCCAGGCGAACCUUCUGUCCUU--------CUGCCCGGCCAUUCAAGUCAUCCAUCUCGGCUUUGGCAG -......((((((((((.-----....(((((..........))))).............--------......(((.......))).......)))).)))))).. ( -22.00, z-score = -1.65, R) >droEre2.scaffold_4929 9391464 107 - 26641161 CCAUCCAGCCGAACCGAAAUACUAUACUCGCCCAUCCAUCCAGGCGAACCUUCUGUCCUUCUGUCCUCCUGCCCGGCCAUCCAAGUCAUCCAUCUCGGCUUUGGCAG .......((((((((((..........(((((..........)))))...........................(((.......))).......)))).)))))).. ( -22.00, z-score = -1.30, R) >droAna3.scaffold_12916 10465326 76 - 16180835 -------------------------------CCAUCCAGCCAGGCCAAAACCUAAACCAUCCAUCCAUCCAAAUGUCCAUCCAAGCCAUCCAUCUCGGAUUUGGCAG -------------------------------.......(((((((.......................................)))((((.....)))).)))).. ( -10.83, z-score = -1.20, R) >consensus CCAUCCAGCCGAACCGAA_____AUACUCGCCCAUCCAUCCAGGCGAACCUUCUGUCCUU________CUGCCCGGCCAUUCAAGUCAUCCAUCUCGGCUUUGGCAG .......((((((((((..........(((((..........)))))...........................(((.......))).......)))).)))))).. (-10.88 = -13.88 + 3.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:58 2011